The accessory helix of complexin functions by stabilizing central helix secondary structure
Figures
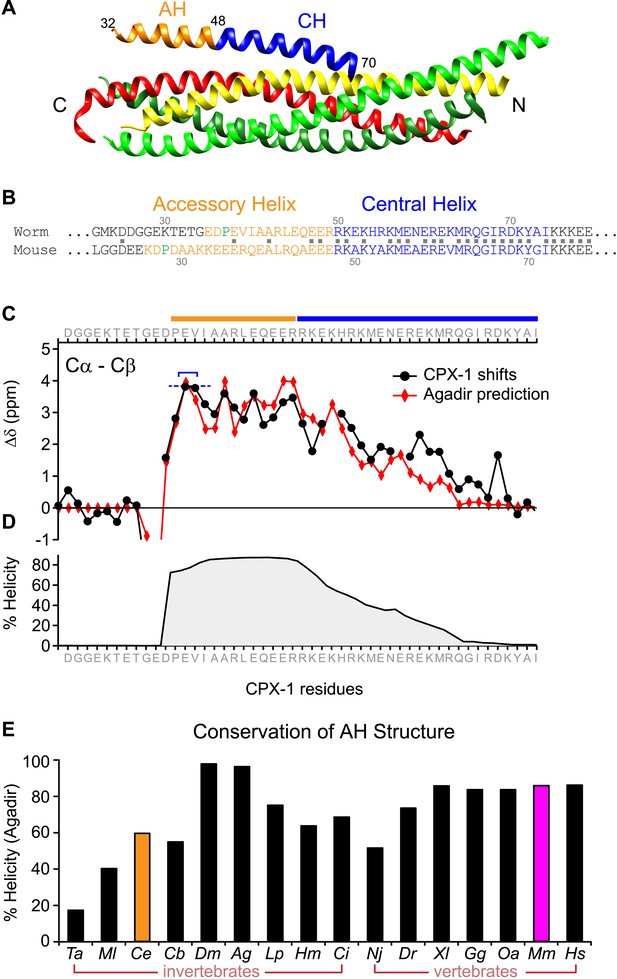
The worm AH domain forms a stable helix and this structure is deeply conserved across phylogeny.
(A) Ribbon diagram of the mammalian complexin-SNARE crystal structure (Chen et al., 2002) using PDB code 1KIL. Cytoplasmic SNARE domains from synaptobrevin (red), Syntaxin (yellow), and SNAP-25 (green). Mouse Cpx1 (residues 26–83) is divided into the accessory helix (AH–orange) and central helix (CH–blue). (B) Sequence alignment of the accessory helix (orange) and central helix (blue) for C. elegans CPX-1 (worm) and M. musculus Cpx1 (mouse). Amino acid identity indicated with gray squares in between the sequences. Helix-breaking prolines indicated in green. (C) Cα-Cβ shifts from a truncated worm CPX-1 peptide missing the C-terminal domain (residues 1–77, black circles) are compared with Agadir predictions for the Cα shifts (red diamonds). The Agadir predicted Cα shift values at residues E38 and V39 were normalized to the experimentally determined Cα-Cβ shift values at those sites (blue bracket), allowing for a comparison of AH and CH shift predictions. (D) Predicted helical state of each residue (Agadir) is shown for CPX-1. (E) Summary of Agadir helix prediction for the AH domain (defined by the average helicity of 18 residues N-terminal to the CH domain) across 16 species: Trichoplax adhaerens Ta, Mnemiopsis leidyi Ml, Caenorhabditis elegans Ce (orange), Caenorhabditis briggsae Cb, Drosophila melanogaster Dm, Anopheles gambiae Ag, Loligo pealei Lp, Hirudo medicinalis Hm, Narke japonica Nj, Ciona intestinalis Ci, Danio rerio Dr, Xenopus laevis Xl, Gallus gallus Gg, Ornithorhynchus anatinus Oa, Mus musculus Mm (pink), Homo sapiens Hs.
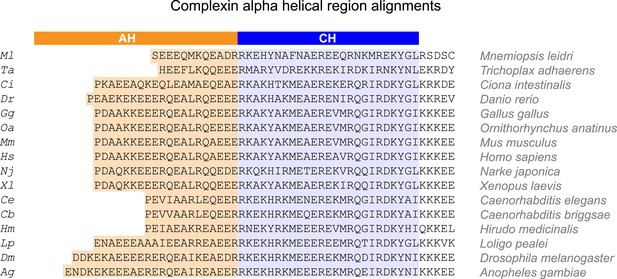
Alpha helical regions of complexin across phylogeny.
Alignments of the alpha helical regions of complexin across 16 species from seven phyla using Clustal Omega multiple sequence alignment. The accessory helix region (orange) was defined based on Agadir prediction of at least 5% helicity. The central helix domain is shaded in blue. Species are indicated on right and by a two letter code used throughout the paper (left). Sequences derived from Ensembl database (Flicek et al., 2014) and aligned using Clustal Omega (EMBL-EBI).

Evolutionary conservation of helicity across >0.5 billion years.
(A) Agadir helicity predictions for human (Hs, blue), ctenophore (Ml, pink), placazoa (Ta, green), and worm (Ce, dashed line) complexin homologs, plotted relative to the first residue of the central helix (position #1). (B). Average domain helicity from (A) using AH (orange, residues −19 to 0) and CH (blue, residues 1 to 25) domains for the four species shown in A. (C) Percent sequence identity relative to human mCpx1 for the AH (orange) and CH (blue) domains for the three invertebrate complexins. Note that the placazoan Trichoplax does not possess a nervous system.
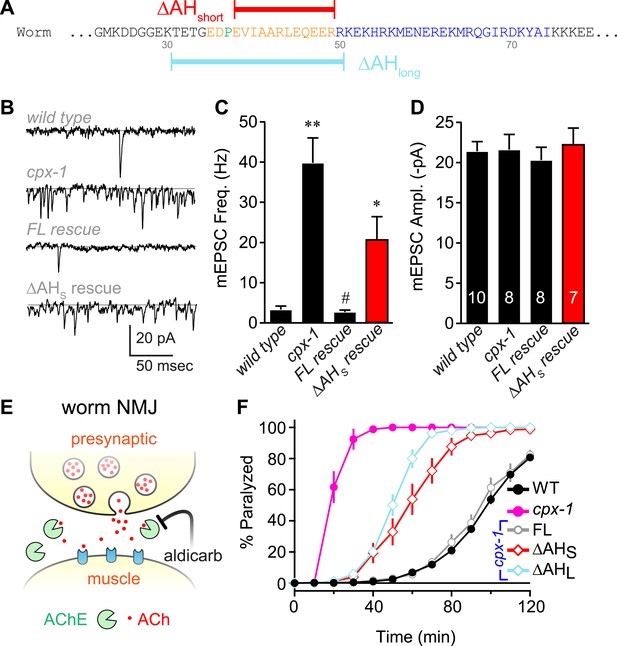
The worm AH contributes to CPX-1 inhibition of spontaneous vesicle fusion.
(A) Two deletions within the worm AH domain were used: ΔAHshort (35–49, red) and ΔAHlong (30–50, aqua). (B) Examples of spontaneous EPSCs in zero external Ca2+ for wild-type, cpx-1, and transgenic animals expressing full-length CPX-1 (FL rescue) and the short AH deletion (ΔAHshort rescue). Average spontaneous EPSC Rate (C) and EPSC amplitude (D) for the genotypes indicated in B. Data are mean ± SEM and the number of independent assays is indicated for each genotype. Using Tukey–Kramer statistics for multiple comparisons, ** denotes significantly different from wild type, # significantly different from cpx-1 but not wild type, * significantly different from both wild type and cpx-1 (p < 0.01). (E) Cartoon of aldicarb acting at the worm cholinergic neuromuscular junction. Acetylcholine (ACh, red) is hydrolyzed by cleft cholinesterases (AChE, green). Aldicarb inhibits AChE causing an elevation in ACh and eventual paralysis depending on the level of exocytosis. (F) Paralysis time course on 1 mM aldicarb for wild-type (black filled circles, n = 36), cpx-1 (pink filled circles, n = 10), full-length rescue CPX-GFP (gray open circles, n = 10), ΔAHshort rescue (red open diamonds, n = 10), and ΔAHlong rescue (aqua open diamonds, n = 10).

Axonal protein abundance for CPX-1 transgenes.
(A) Schematic of a worm depicting the region imaged for axonal expression (pink box). All rescuing transgenes were quantified by imaging the C-terminal GFP tag in single axons of immobilized intact animals. (B) Representative confocal image of CPX-1::GFP fluorescence in a dorsal cord axon of an intact animal. Scale bar is 5 microns. (C) Average axonal fluorescence values for transgenic animals used in this study. Fluorescence was background-subtracted and averaged for at least 20 animals for each strain (number indicated in the bars). All data were normalized to a full-length wild-type CPX-1::GFP rescue strain and plotted as mean ± SEM. The blue dotted lines represent the range of expression found to rescue aldicarb sensitivity based on prior studies using a previously described axonal expression quantification protocol (Wragg et al., 2013).
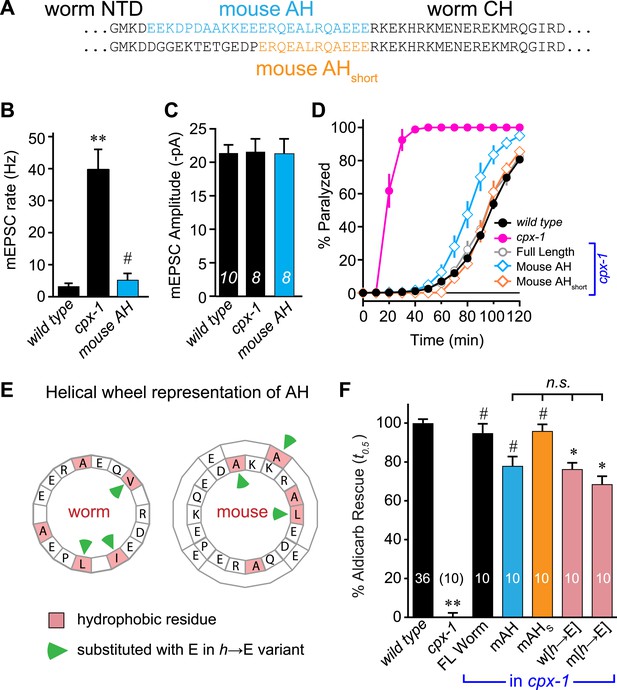
Mouse AH functions in worm CPX-1 and neither domain requires hydrophobic residues.
(A) Two chimeric CPX-1 constructs substituting worm AH with the mouse AH. The long form (mouse AH) substitutes worm residues 26–49 with mouse residues 24–47 whereas the short form (mouse AHshort) substitutes worm residues 38–49 with mouse residues 36–47. Average spontaneous mEPSC rates (B) and amplitudes (C) for wild-type, cpx-1, and transgenic rescue of cpx-1 expressing the mouse AH chimera (mouse AH, blue). (D) Average paralysis time course in 1 mM aldicarb for wild-type (black filled circles), cpx-1 (pink filled circles), full-length rescue (gray open circles), mouse AH rescue (blue open diamonds), and mouse AHshort rescue (orange open diamonds). (E) Worm and mouse AH domain residues shown on a helical wheel diagram with hydrophobic residues indicated in red and hydrophobic substitutions with glutamate indicated with green arrow heads. (F) Percent rescue of wild-type paralysis kinetics using the time to 50% paralysis (t0.5) for the five genotypes shown in D as well as a worm CPX-1 variant with V39E, I40E, L44E substitutions (w[h→E]) and a mouse AHshort variant with A40E, L41E, A44E substitutions (m[h→E]). Note that the residue positions are labeled based on their location in CPX-1 and mCpx1 respectively. Data are mean ± SEM with sample sizes as indicated on the bar graphs. Using Tukey–Kramer statistics for multiple comparisons, ** denotes significantly different from wild type, # significantly different from cpx-1 but not wild type, * significantly different from both wild type and cpx-1 (p < 0.01), n.s. is not significant.
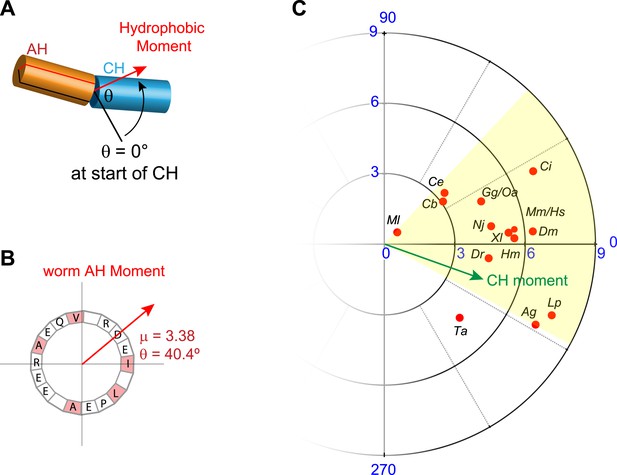
Conservation of hydrophobic moments of complexin.
(A) The AH hydrophobic moment was computed with angular orientation relative to the first residue of the CH. (B) Example of a hydrophobic moment for a particular AH (worm). (C) Polar plot of the hydrophobic moments for 16 species (red) and rescuing constructs (turquoise). All species besides Trychoplax cluster in a small range of orientations (yellow). The moment of the worm CH is shown in green. The 16 species shown are Trichoplax adhaerens Ta, Mnemiopsis leidri Ml, Caenorhabditis elegans Ce, Caenorhabditis briggsae Cb, Drosophila melanogaster Dm, Anopheles gambiae Ag, Loligo pealei Lp, Hirudo medicinalis Hm, Narke japonica Nj, Ciona intestinalis Ci, Danio rerio Dr, Xenopus laevis Xl, Gallus gallus Gg, Ornithorhynchus anatinus Oa, Mus musculus Mm, and Homo sapiens Hs.
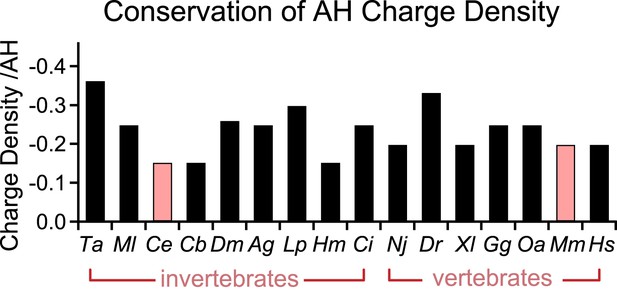
Conservation of charge density in the AH domain.
The helical region of the AH domain was defined using Agadir with a criterion of >5% predicted helicity for a continuous region N-terminal to the CH domain. The net formal charge within this stretch was divided by the number of residues to calculate a charge density and plotted as charge density/AH for 16 species. C. elegans and M. musculus (pink) are the species investigated in this study. The 16 species shown are Trichoplax adhaerens Ta, Mnemiopsis leidri Ml, Caenorhabditis elegans Ce, Caenorhabditis briggsae Cb, Drosophila melanogaster Dm, Anopheles gambiae Ag, Loligo pealei Lp, Hirudo medicinalis Hm, Narke japonica Nj, Ciona intestinalis Ci, Danio rerio Dr, Xenopus laevis Xl, Gallus gallus Gg, Ornithorhynchus anatinus Oa, Mus musculus Mm, and Homo sapiens Hs.
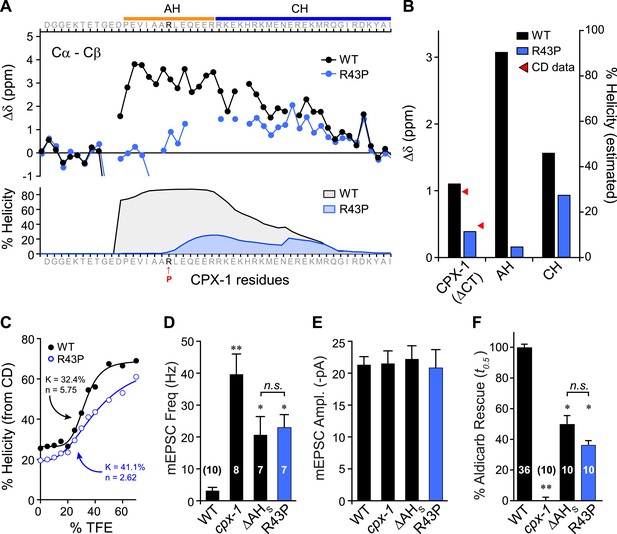
Disrupting AH helix stability impairs CPX-1 inhibitory function.
(A) NMR derived Cα-Cβ shifts from either wild-type (black) or R43P (blue) worm CPX-1 peptide missing the C-terminal domain. R43P is indicated in red. Below, the predicted helical content using Agadir for wild-type (black), and R43P complexin (blue). (B) Average secondary chemical shift for wild-type (black) and R43P (blue) complexin either over the entire peptide (residues 1–77, left), AH domain (37–49, middle), or CH domain (50–74, right). The average helical content was estimated by dividing the chemical shift by 3.4 (average shift of 100% helical peptide). The helical content was also measured by CD spectroscopy (red arrowheads). (C) Helical content for wild type (black) and R43P (blue) complexin was measured by CD spectroscopy for increasing concentrations of 2,2,2-trifluoroethanol (TFE). The resulting dose–response data was fit to a simple equilibrium binding curve with equilibrium constants and Hill coefficients indicated on the graph. Average spontaneous EPSC Rate (D) and EPSC amplitude (E) for wild-type, cpx-1, and either the ΔAHshort or R43P rescuing transgene expressed in cpx-1 as indicated. (F) Sensitivity to aldicarb was quantified by the average time to 50% paralysis and then normalized to wild-type and cpx-1 mutant animals. On this scale, rescue with ΔAHshort or R43P variants of CPX-1 partially restored wild-type aldicarb sensitivity. Data are mean ± SEM and the number of independent assays is indicated for each genotype. Using Tukey–Kramer statistics for multiple comparisons, ** denotes significantly different from wild type, * significantly different from both wild type and cpx-1 (p < 0.01), n.s. is not significant.
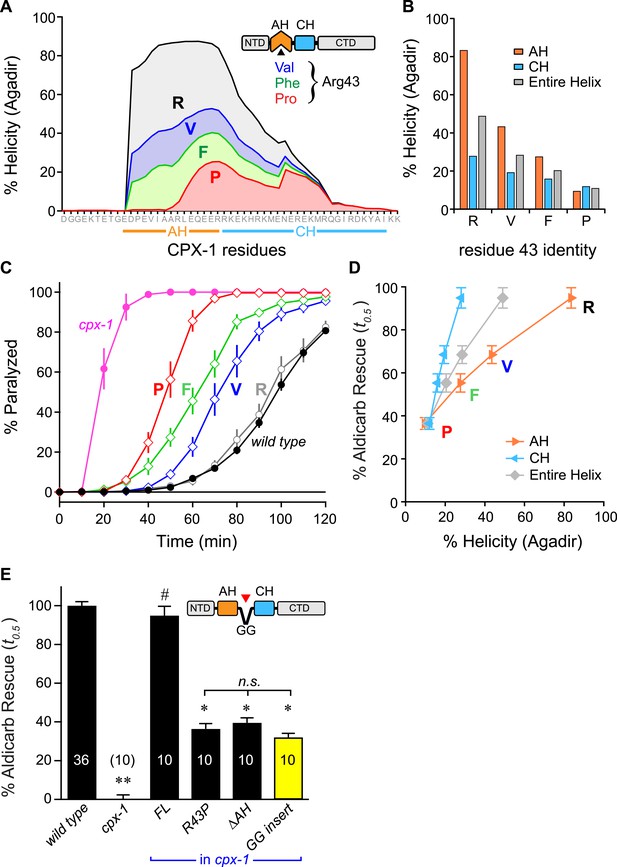
Stability of the AH and its propagation into the CH domain are required for CPX-1 inhibitory function.
(A) AH and CH helical content based on Agadir predictions is plotted for wild-type CPX-1 and three variants with a single substitution at residue 43 as indicated. (B) Average helical content of the AH domain (orange), CH domain (blue), or entire helical region (gray) for four residues in position 43: Arg, Val, Phe, and Pro. (C) Average paralysis time course on one millimolar aldicarb for wild-type (black filled circles) and cpx-1 animals (pink filled circles), as well as four rescuing CPX-1 transgenes: wild type CPX-1 (R, gray open circles), R43V (V, blue diamonds), R43F (F, green diamonds), and R43P (P, red diamonds). (D) Normalized rescue of the t0.5 for paralysis for each of the four transgenic rescue strains is plotted vs the predicted helicity for either the AH domain (orange), the CH domain (blue), or the entire helical region (gray). (E) Normalized rescue of aldicarb sensitivity (t0.5 ) for wild-type and cpx-1 animals as well as three transgenic rescue strains: full-length wild-type CPX-1 (FL), R43P, the long AH deletion (ΔAH), and the GG insert in between the AH and CH domains (GG insert). Data are mean ± SEM with n = 10 experiments for all strains except wild type (n = 36). **p < 0.01 different from wild-type, # is p < 0.01 different from cpx-1 but not wild type. *p < 0.01 different from wild type and cpx-1 animals. Significance was determined by Tukey–Kramer method.
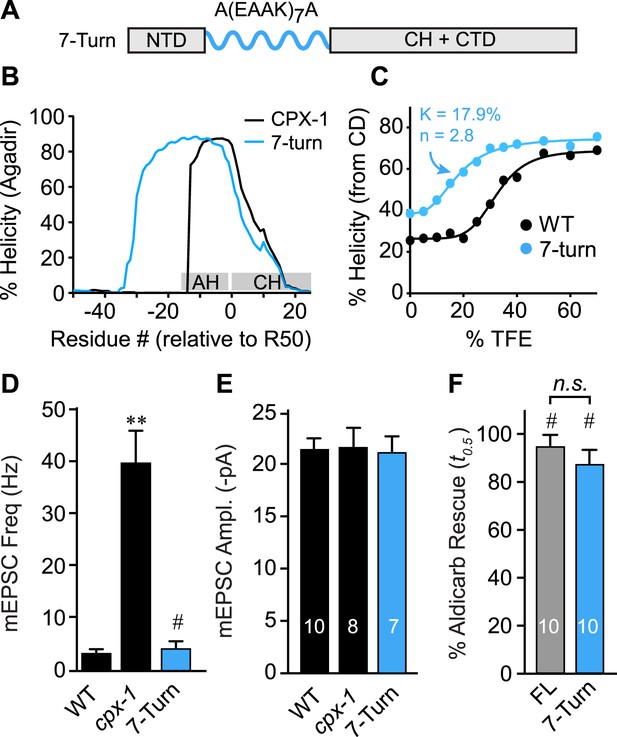
The AH domain can be functionally replaced by a non-native helix.
(A) Schematic of the helix substitution strategy. A(EAAK)7A sequences were substituted for residues 26–49 in the worm AH domain. (B) Agadir prediction for helical stability of the 7-turn helix motif compared to wild-type AH. (C) Helical content for the 7-turn construct was measured by CD spectroscopy, and the TFE dose–response data were fit as in Figure 4. Average spontaneous EPSC Rate (D) and EPSC amplitude (E) for wild-type, cpx-1, and the 7-turn rescuing transgene expressed in cpx-1 as indicated. (F) Sensitivity to aldicarb was quantified by monitoring the average time to 50% paralysis normalized to wild-type and cpx-1 mutant animals, and plotted for full-length wild-type CPX-1 (FL), 5-turn substitution (5 Turn), and 7-turn substitution variants expressed in cpx-1 mutants. Data are mean ± SEM and the number of independent assays is indicated for each genotype. Using Tukey–Kramer statistics for multiple comparisons, ** denotes significantly different from wild type, # significantly different from cpx-1 but not wild type, * significantly different from both wild type and cpx-1 (p < 0.01), n.s. is not significant.
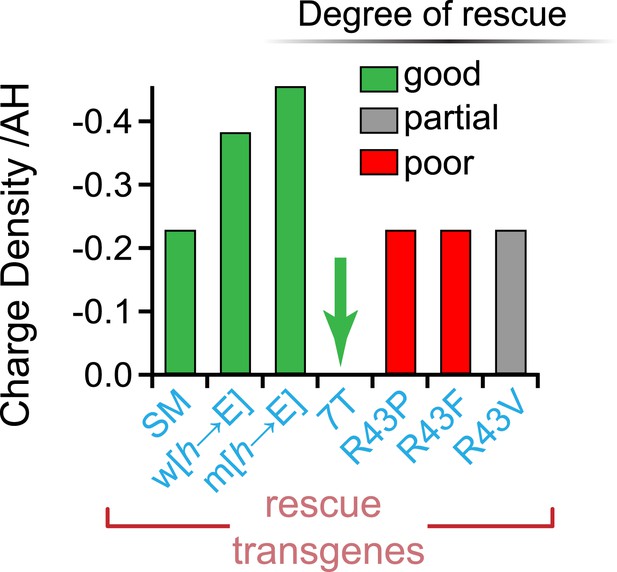
No correlation between charge density and function.
Charge density of the AH domain in seven rescuing CPX-1 variants. Green indicates nearly complete rescue, gray is partial, and red is poor rescue as measured by aldicarb sensitivity. The CPX-1 variants are mouse AHshort (SM), worm AH with hydrophobic to glutamates (w[h→E]), mouse AH with hydrophobic to glutamates (m[h→E]), 7-turn helix substitution (7T), as well as the R43P, R43F, and R43V substitutions.
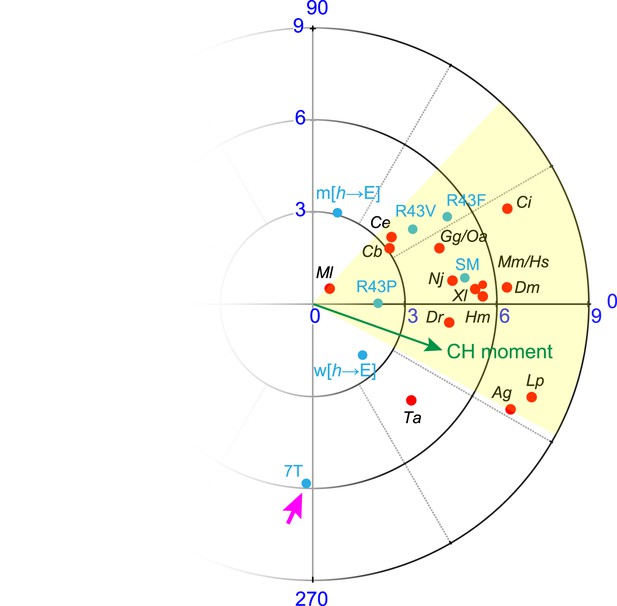
Hydrophobic moments of functional and nonfunctional CPX transgenes.
Polar plot of hydrophobic moments for the AH domain of CPX-1 variants. The CPX-1 variants (blue) are mouse AHshort (SM), worm AH with hydrophobic to glutamates (w[h→E]), mouse AH with hydrophobic to glutamates (m[h→E]), 7-turn helix substitution (7T), as well as the R43P, R43F, and R43V substitutions. The moment of the worm CH is shown in green. 7T construct designated with a pink arrow. The hydrophobic moments of 16 species from Figure 3—figure supplement 1 are included for comparison.





