Ngn1 inhibits astrogliogenesis through induction of miR-9 during neuronal fate specification
Figures
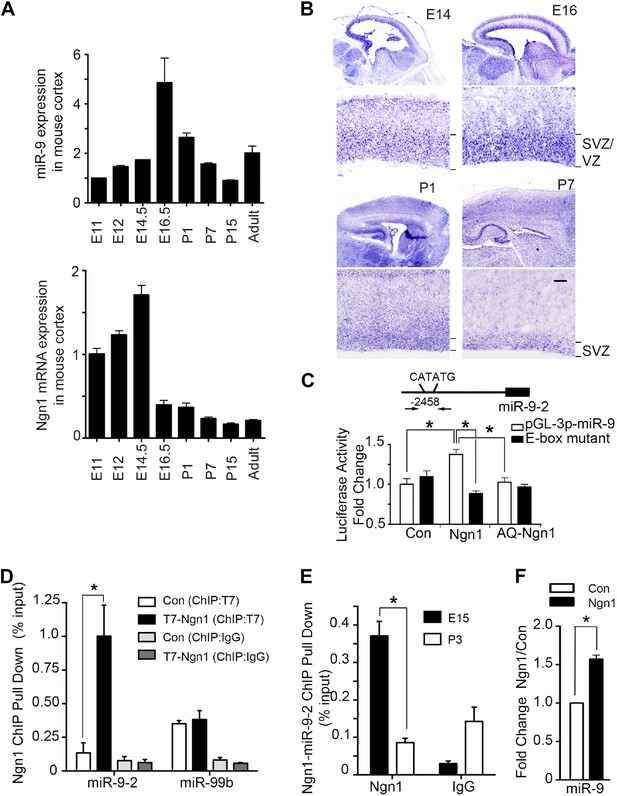
Neurogenin 1 (Ngn1) directly regulated miR-9 expression.
(A) Temporal expression of miR-9 and Ngn1 in the developing and adult mouse cortex. Upper panel: Taqman quantitative PCR (qPCR) analysis of endogenous miR-9 expression levels. Data were normalized to the loading control U6 non-coding RNA. Lower panel: qRT-PCR of endogenous Ngn1 mRNA levels. Data were normalized to loading control Gapdh. Gapdh, glyceraldehyde-3-phosphate dehydrogenase. (B) Expression analysis of miR-9 in the prenatal and neonatal mouse cortex by in situ hybridization (ISH). The mature miR-9 was extensively expressed in the VZ/SVZ progenitors in developing cortex. VZ, ventricular zone. SVZ, subventricular zone. Scale bar: 100 μm. (C) Ngn1 up-regulated the activity of the miR-9-2 promoter-driven luciferase reporter but not Ngn1 binding site mutant Ngn1 (AQ-Ngn1). (D) ChIP-qPCR analysis showed Ngn1 bound to miR-9-2 but not miR-99b promoter region in mouse E11 cortical NPCs overexpressing T7-tagged Ngn1. Data was presented as percentage pull down (IP using T7 antibody) comparing to the input. Negative control IgG pull down was also shown (*p < 0.05, Mann–Whitney test). (E) ChIP-qPCR of endogenous Ngn1 associated with miR-9-2 promoter in E15 and in P3 mouse cortexes. Data was presented as percentage pull down (IP using Ngn1 antibody) comparing to the input. Negative control IgG pull down was also shown (*p < 0.05, Mann–Whitney test). (F) Taqman qPCR analysis of the expression level of miR-9 in mouse NPCs in the presence of T7-tagged Ngn1 (*p < 0.02, Mann–Whitney test).
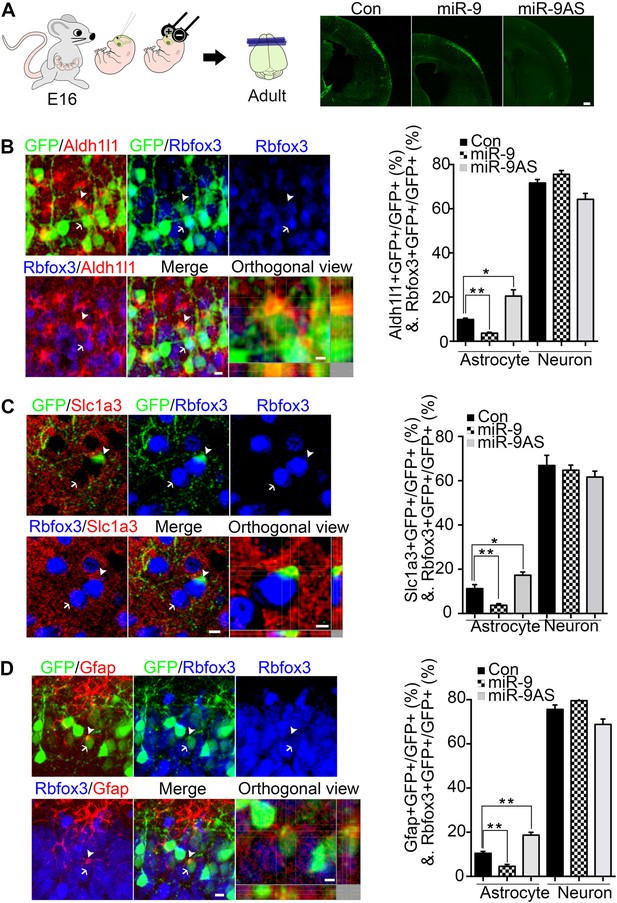
miR-9 inhibited astrogliogenesis in vivo.
(A) Schematic representation of in utero injection of miR-9 constructs followed by electroporation into cortical progenitors resided at the ventricular surface at E16. The right panels showed that cells electroporated at E16 (green) with each condition all migrated to proper cortical layers without overt migration defects. Scale bar: 100 μm. (B–D) Left panels showed examples of astrocyte marker+ or neuronal marker+ cells. The arrow pointed Rbfox3+(NeuN) cortical neuron (green and blue). The arrowhead pointed astrocyte marker+ astrocyte (green and red). Scale bar: 10 μm. The arrowhead pointed astrocyte was shown at greater magnification in the last left panel (confocal montage with orthogonal views taken at the center of the cell, scale bar: 5 μm). Right panels showed that overexpression of miR-9 in vivo dramatically attenuated astrogliogenesis, whereas knockdown of miR-9 had the opposite effect (**p < 0.01, *p < 0.5, Mann–Whitney test). Aldh1l1, aldehyde dehydrogenase 1 family, member L1; Slc1a3 (EAAT1), solute carrier family 1 (sodium-dependent glutamate/aspartate transporter 1), member 3; Gfap, glial fibrillary acidic protein.
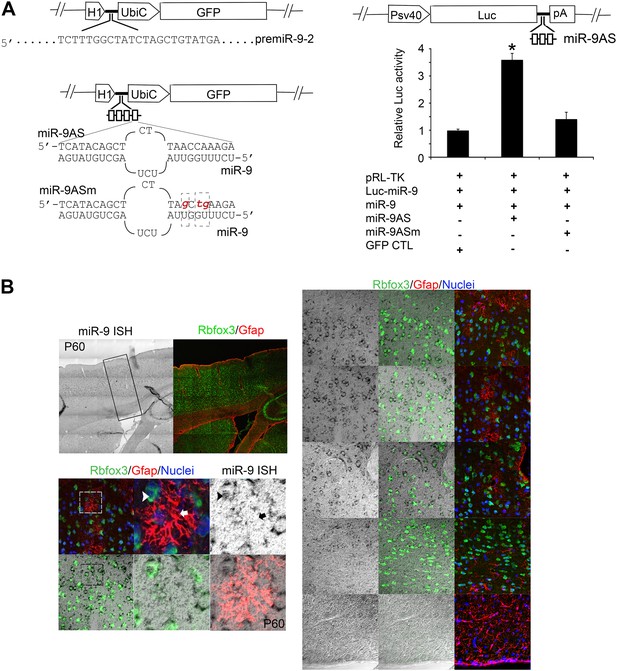
(A) Upper left panels showed construction of miR-9 overexpression plasmid (miR-9) by inserting pre-miR-9-2 and its flanking sequences to a modified lentiviral plasmid FG12.
Lower left panels showed the construction of miR-9 knockdown lentiviral vector (miR-9AS) and knockdown control (miR-9ASm). Four miR-9 bulged antisense sequences were inserted immediately downstream of the H1 promoter of the modified-FG12 vector. H1, H1 promoter. UbiC, ubiquitin promoter. miR-9AS mutant control, miR-9Sm was constructed by modifying 3-nt in miR-9 seed binding region. Right panels showed the luciferase assay of miR-9 knockdown efficiency. Upper right panel showed the construction of miR-9 luciferase assay plasmid. Three miR-9 bulged antisense repeats were inserted immediately downstream of a firefly luciferase gene. Lower right panel showed relative fold changes of luciferase activity. miR-9 luciferase construct was co-transfected into HEK239T cells with miR-9 and/or miR-9AS. A Renilla luciferase plasmid pRL-TK was used as an internal control. The empty vector FG12 (con) and the miR-9 binding site mutant miR-9AS (miR-9ASm) were used as negative controls. *p < 0.05, Wilcoxon-Mann-Whitney test. (B) Endogenous miR-9 was expressed in neocortical neurons, but not in astrocytes in adult cortex. miR-9 ISH combined with Rbfox3 (NeuN) and Gfap double immunostaining in the upper left panels were shown at greater magnification in the right panels. The lower left panels showed miR-9 positive neuron that overlaps with a neuronal maker, Rbfox3 (green, arrowhead), but not Gfap positive astrocyte (red, arrow).
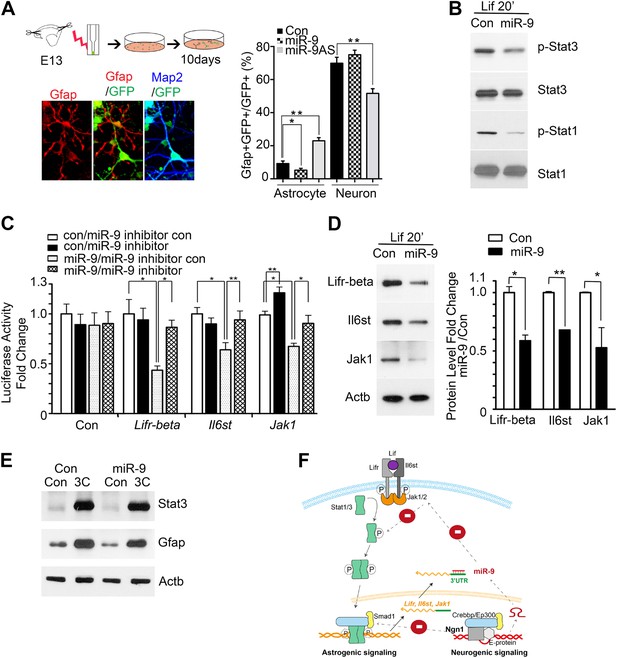
miR-9 inhibited astrogliogenesis via targeting three components of Jak-Stat pathway.
(A) Upper right panels: schematic representation of in vitro delivery of miR-9 constructs into cortical progenitors by electroporation. Lower left panels showed examples of astrocyte marker Gfap+ or neuronal marker Map2+ cells. Right panel showed overexpression of miR-9 significantly reduced the number of astrocytes 10 days after electroporation, whereas knockdown of miR-9 dramatically promoted astrogliogenesis (**p < 0.01, *p < 0.5, Mann–Whitney test). Overexpression of miR-9 in NPCs did not increase the number of neurons. Knockdown of miR-9 reduced the number of Map2+ neurons. Map2, Microtubule-Associated Protein 2. (B) Transfection of mouse NPCs with exogenous miR-9 duplex blocked phosphorylation of Stat1/3 without altering their protein levels. (C) Luciferase activity of Lifr-beta, Il6st (gp130), and Jak1 3′ UTR luciferase reporter in the presence of different combinations of control (con), miR-9, miR-9 inhibitor con, and miR-9 inhibitor in mouse NPCs. (D) miR-9 inhibited protein levels of Jak-Stat signaling components in NPCs transfected with control or miR-9 duplex. Right panels: western blotting densitometry analysis of protein level changes. Actb (β-actin) serves as the loading control. (E) A constitutively active form of Stat3, Stat3C bypassed the effect of miR-9 inhibition on astrocyte differentiation. (F) Schematic representation of Ngn1-regulated miR-9 signaling that modulates Stat1/3 phosphorylation to control cell fate specification. Ngn1 up-regulates miR-9 expression during neurogenesis. miR-9 reduces protein levels of Lifr-beta, Il6st, and Jak1 of Jak-Stat signaling pathway by targeting their 3′ UTRs, which in turn abolish Stat1/3 phosphorylation to suppress astrogliogenesis. Crebbp: CREB binding protein, Smad1: Mothers Against DPP Homolog 1 (Drosophila), Ep300: E1A Binding Protein p300, E-protein: ubiquitous basic-Helix-Loop-Helix proteins, such as E12 or E47. The dotted arrows show the inhibition regulations.
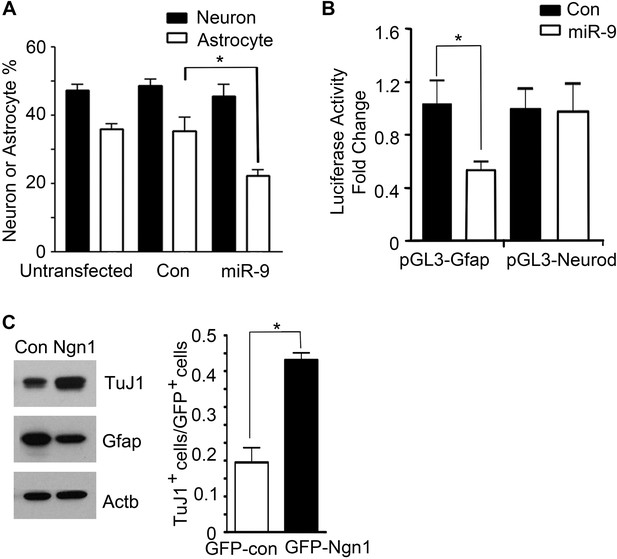
miR-9 and Ngn1 inhibited astrogliogenesis.
(A) Transfection of mouse E11 cortical NPCs with exogenous miR-9 duplex significantly reduced the number of Gfap+ astrocytes in vitro. (B) miR-9 suppressed activation of glial-specific Gfap promoter, with little effect on the neurogenic promoter Neurod1. (C) Ngn1 promoted neurogenesis and suppresses astrogliogenesis. Western blotting analysis showed that overexpression of Ngn1 in mouse E11 cortical NPC promoted Tuj1 expression, while reduced Gfap+ expression (left panels). Tuj1, beta III tubulin, a neuronal marker. Right panel showed overexpression of Ngn1 in mouse E11 cortical NPC promoted neurogenesis (Tuj1+ cells).
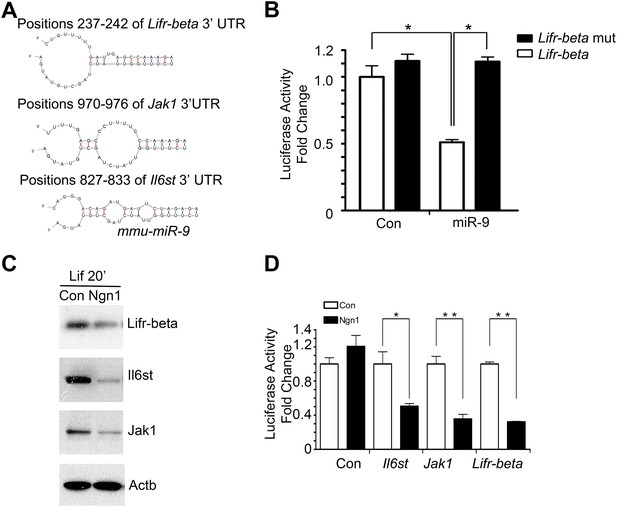
miR-9 targeted Jak-Stat signaling pathway.
(A) Predicted duplex formation between mouse Lifr-beta, Il6st, and Jak1 3′ UTR (top) and miR-9 (bottom). (B) Luciferase activity of wild type Lifr-beta 3′ UTR and miR-9 binding region mutant Lifr-beta 3′ UTR reporter genes with or without miR-9 duplex in mouse E11 cortical NPCs (*p < 0.0001, Wilcoxon-Mann-Whitney test). (C, D) Ngn1 suppressed the expression of three major components of Jak-Stat signaling pathway. (C) Western blot analysis of Ngn1 suppressed the protein levels of Lifr-beta, Il6st, and Jak1. Actb served as the loading control. (D) Overexpression of Ngn1 decreased the activity of luciferase reporter fused to 3′ UTRs of Il6st, Lifr-beta, and Jak1 mRNAs. (*p < 0.05, **p < 0.005, Wilcoxon-Mann-Whitney test).





