Species-wide whole genome sequencing reveals historical global spread and recent local persistence in Shigella flexneri
Figures
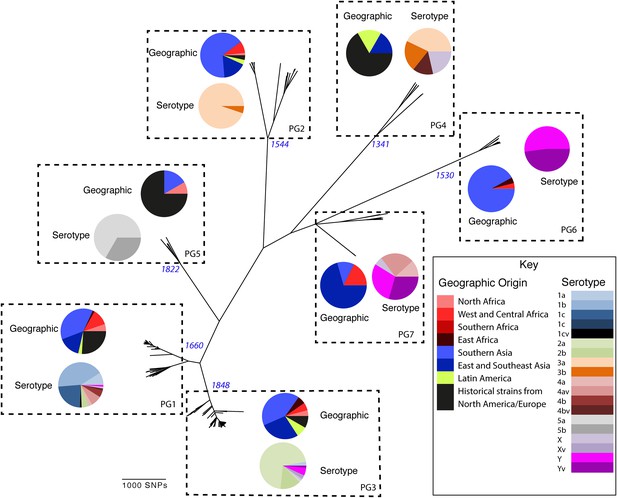
Maximum likelihood phylogeny for Shigella flexneri isolates including serotypes 1–5, X and Y produced from the results of mapping sequence reads against the genome of S. flexneri 2a strain 301, with recombination removed.
Phylogenetic groups (PGs) determined by Bayesian analysis of population structure clustering are boxed within dotted lines, with the geographic and serotype composition of isolates in each PG being inlaid as pie charts.
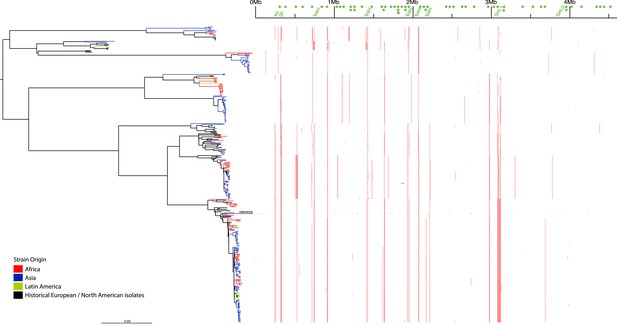
Location of segments detected as recombinant.
Blue indicates a likely recombination within an individual isolate while red indicated recombination common to multiple isolates. Green text at the top indicates mobile elements determined by a manual examination of the reference S. flexneri strain 301 genome.
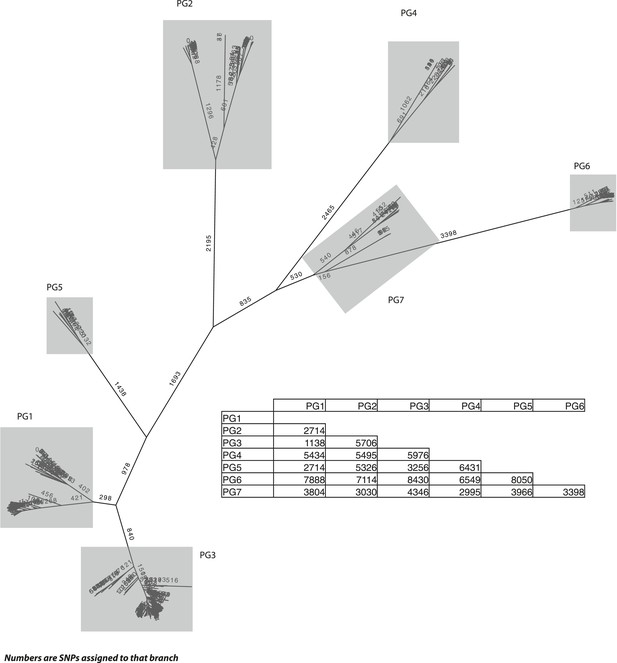
S. flexneri species tree, with the number of single nucleotide polymorphisms (SNPs) per branch.
The SNP tree uses the same alignment as in Figure 1, but is constructed from the SNPs that can be assigned to each branch. The ancestral states were reconstructed using ACTRAN. Insert—a table showing the number of SNPs between the most recent common ancestor (MRCA) of each of the PGs identified.
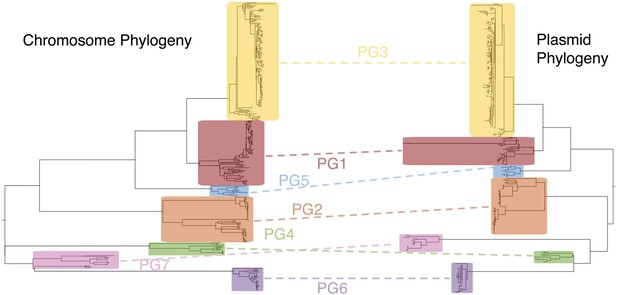
Co-evolutionary relationships of the S. flexneri genome and virulence plasmid (VP).
A maximum likelihood phylogeny of the S. flexneri chromosome (left) is shown adjacent to one of the VP (right). Collared blocks and labels enclose independently identified BAPs clusters for sequence alignments of the chromosome and VP. Dotted lines indicate groups of isolates shared between clusters in phylogeny.
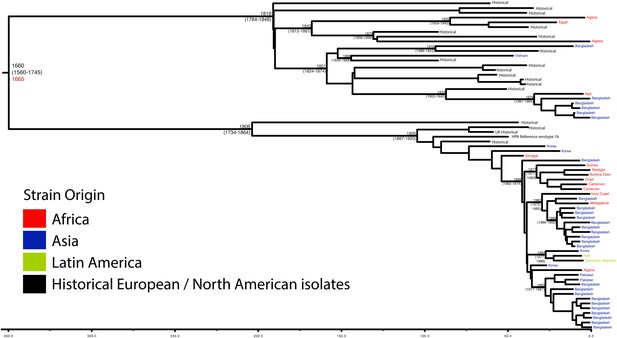
Maximum Clade Credibility trees generated using Bayesian evolutionary analysis by sampling trees (BEAST) for PG 1.
Dates of MRCA are shown overlying internal nodes followed by 95% HPD in parentheses. Tips display the country of origin for each isolate (where available), coloured by region while the date given in red at the base of each group is the MRCA date obtained from the software Path-O-Gen, calculated based on the root-to-tip distance. The horizontal scale is in the unit of years in the past.
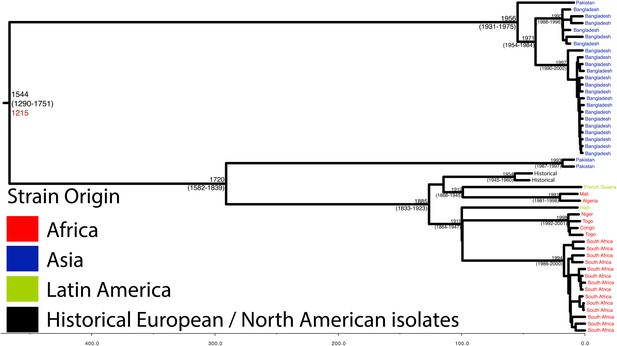
Maximum Clade Credibility trees generated using BEAST for PG 2.
Dates of MRCA are shown overlying internal nodes followed by 95% HPD in parentheses. Tips display the country of origin for each isolate (where available), coloured by region while the date given in red at the base of each group is the MRCA date obtained from the software Path-O-Gen, calculated based on the root-to-tip distance. The horizontal scale is in the unit of years in the past.
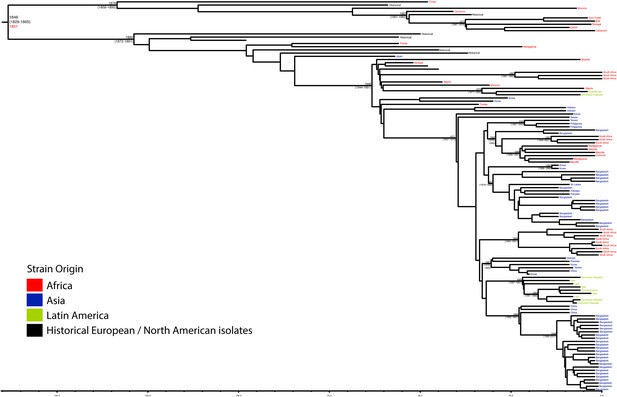
Maximum Clade Credibility trees generated using BEAST for PG 3.
Dates of MRCA are shown overlying internal nodes followed by 95% HPD in parentheses. Tips display the country of origin for each isolate (where available), coloured by region while the date given in red at the base of each group is the MRCA date obtained from the software Path-O-Gen, calculated based on the root-to-tip distance. The horizontal scale is in the unit of years in the past.
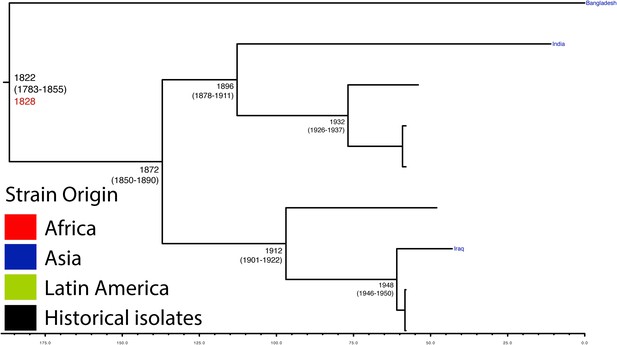
Maximum Clade Credibility trees generated using BEAST for PG 5.
Dates of MRCA are shown overlying internal nodes followed by 95% HPD in parentheses. Tips display the country of origin for each isolate (where available), coloured by region while the date given in red at the base of each group is the MRCA date obtained from the software Path-O-Gen, calculated based on the root-to-tip distance. The horizontal scale is in the unit of years in the past.
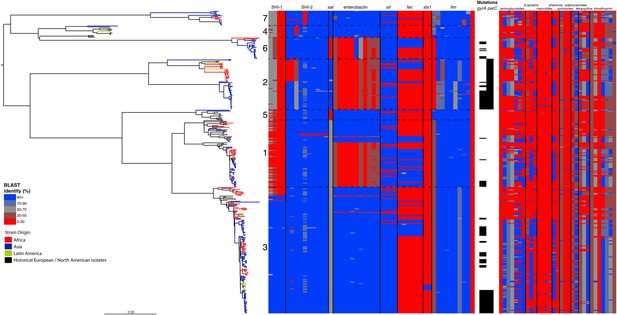
Correlation of isolate phylogeny with pathogenicity and antimicrobial resistance (AMR) determinants.
The midpoint-rooted maximum likelihood phylogenetic tree shows PGs, with tips and terminal branches collared by continent of origin. Tracks adjacent to each isolate show the percentage BLAST identity of the best hit in the sample assembly against key virulence and AMR determinants. Isolates with mutations in the gyr/par genes have black bars in the relevant tracks. The virulence determinants shown are SHI-1 (pic, sigA, set1AB), SHI-2 (shiABCDE, iucABCD, iutA), sat, enterobactin (entABECFD, fepABCDG), sitABCD, fecEDCBAR, stx1ab, fimZBCHGFDEAY, and the AMR genes are aac(3)-II, aadA1, aadA2, aadA5, strA, strB, sat1 (aminioglycosides), blaCTX-M-24, blaOXA-1, blaTEM-1, (β-lactams) ermB, msrE, mphA, mphE, (macrolides) catA1, catB1, (phenicols) qacEΔ1, qnrS1, (quinolones) qepA, sul1, sul2 (sulphonamides) tetA(A), tetA(D), tetA(B) (tetracyclin), dfrA17, dfrA3b, dfrA1, dfrA5, dfrA14 and dfrA8 (trimethoprim).
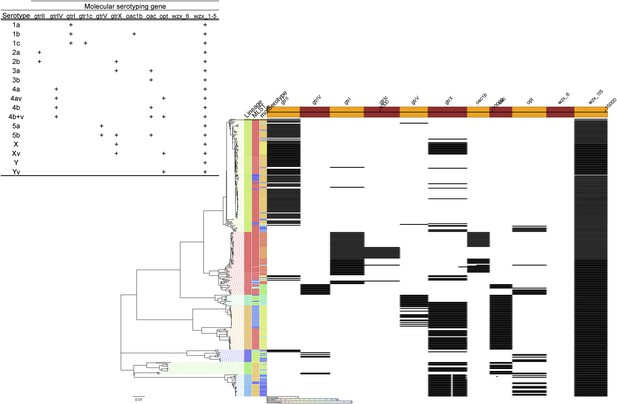
Results of molecular serotyping, displaying the distribution of MLST, molecular serotype, and the distribution of defining genes (according to key, top left) among isolates.
https://doi.org/10.7554/eLife.07335.012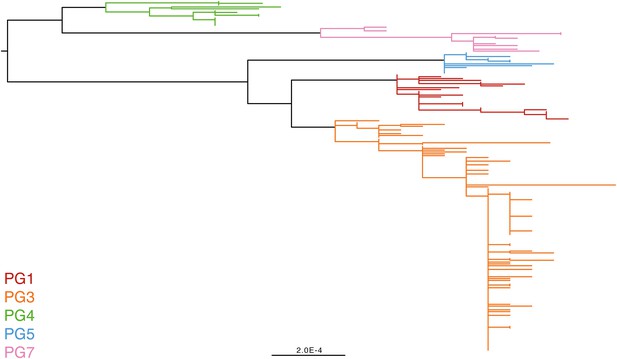
Maximum likelihood phylogeny of an alignment of the concatenated nucleotide sequences the enterobactin locus of 13 genes (34,732 NT; containing entABCDEFS, fepABCDG and fes).
The tree is drawn using PhyML, with a GTR model and contains 191 strains. Isolate labels are collared according to whole-genome based PG definition.
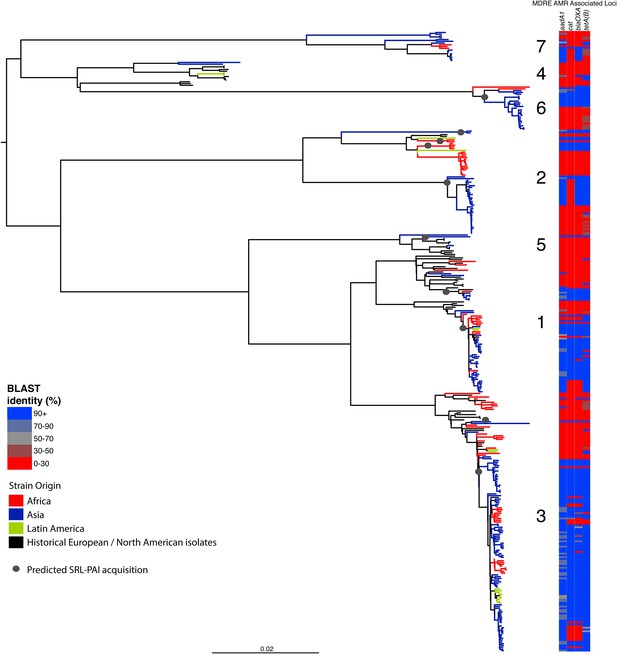
Correlation of isolate phylogeny with AMR determinants, showing only the SRL-MDRE-associated loci aadA1, blaOXA-1, cat and tetA(B).
Grey circles indicate branches where acquisition events are predicted to have taken place.
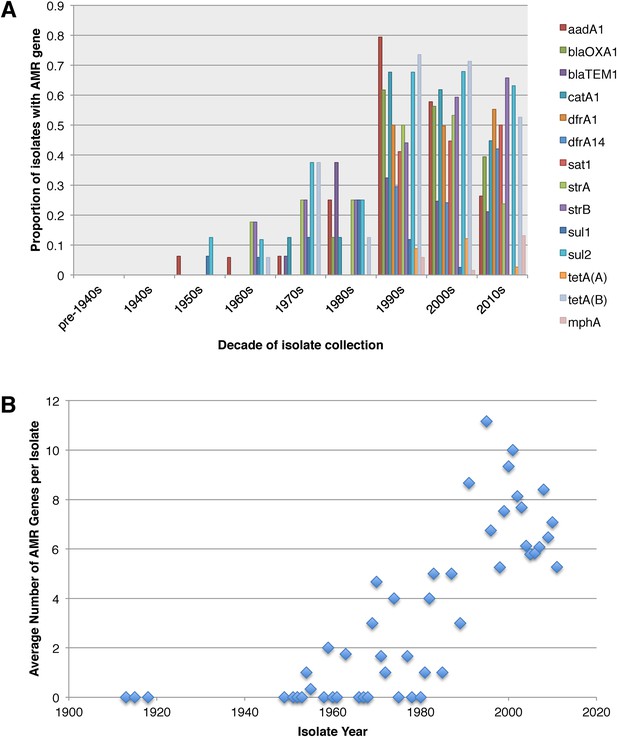
Graphs showing the pattern of AMR presence within our dataset.
(A) Graph showing the proportion of isolates from each decade that contain the AMR genes. (B) Graph showing the average number of resistance genes found in each isolate collected, by year.
Additional files
-
Supplementary file 1
Table containing strain information, accession numbers for the strains used in this study along with the blast identities for the virulence and AMR genes displayed in Figure 2.
- https://doi.org/10.7554/eLife.07335.016





