The Ku subunit of telomerase binds Sir4 to recruit telomerase to lengthen telomeres in S. cerevisiae
Figures
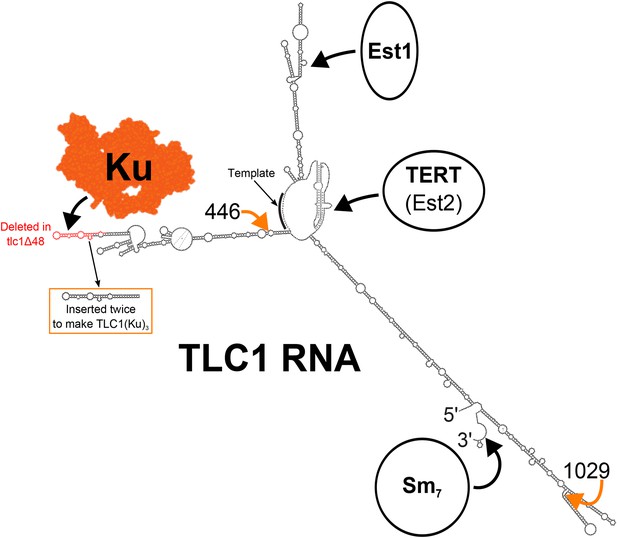
Secondary structure model of yeast telomerase RNA.
The 48 nucleotides deleted in the tlc1Δ48 allele are highlighted in red. The 74-nucleotide hairpin shown in the orange box was inserted at positions 446 and 1029 (indicated by the orange arrows) to create TLC1(Ku)3. The TLC1 secondary structure shown is based on previously published models of the core (Niederer and Zappulla, 2015) and of the arms (Dandjinou et al., 2004; Zappulla and Cech, 2004), while the Ku crystal structure shown is that of the human Ku70/80 complex (Walker et al., 2001).
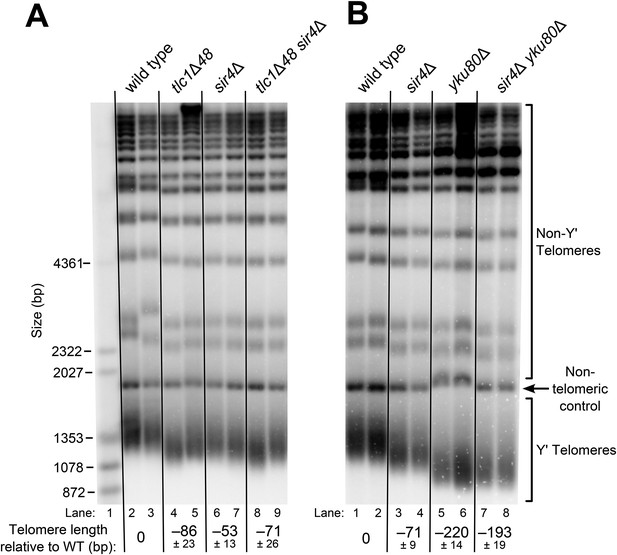
SIR4, Ku, and the Ku-binding site in TLC1 are in the same telomere-lengthening pathway.
(A) Deleting SIR4 in tlc1Δ48 cells does not cause further telomere shortening. A tlc1Δ pTLC1-URA3 strain and an isogenic sir4Δ strain were transformed with CEN plasmids expressing either TLC1 or tlc1Δ48, and then the pTLC1-URA3 cover plasmid was shuffled out. The cells were serially re-streaked five times, and genomic DNA was isolated and analyzed by Southern blotting. The Southern blot was probed for telomeric sequence and for a 1621-bp non-telomeric XhoI restriction fragment from chromosome IV (‘non-telomeric control’) used as a relative-mobility control. Pairs of lanes represent independent transformants. Changes in telomere length were quantitated using the Y′ telomere bands as described in ‘Materials and methods’. Telomere lengths calculated from the two sets of replicates shown were averaged with telomere lengths from four other sets of replicate samples from similar experiments to give the numbers shown, ± standard deviation. The numbers shown here are the same as those in Table 1. (B) Deleting SIR4 in yku80Δ cells does not cause further telomere shortening. A SIR4/sir4Δ YKU80/yku80Δ diploid strain was sporulated, and tetrads were dissected. The haploid spores of a tetratype tetrad were serially re-streaked three times on plates to equilibrate telomere length before Southern blot analysis. The pairs of lanes on the blot shown are different colonies from streak-outs of the haploid spores. Telomere lengths calculated from the two sets of replicates shown were averaged with telomere lengths from a third set of replicate samples to give the numbers shown, ± the standard deviation.
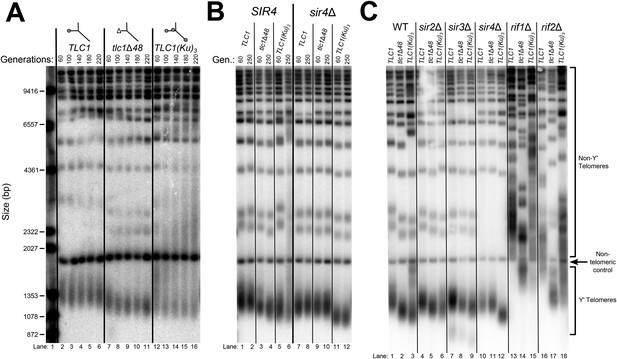
TLC1-bound Ku requires SIR4 to promote telomere lengthening.
(A) TLC1(Ku)3, a TLC1 RNA containing two extra Ku-binding sites, causes both telomere hyper-lengthening and shortening. This experiment was performed as described in Figure 2A, except a tlc1Δ pTLC1-LYS2 rad52Δ strain was used. Additionally, instead of passaging cells on plates, single colonies were inoculated to liquid cultures, which were then serially passaged and harvested at various points throughout the passaging process. (B) TLC1(Ku)3 does not cause telomere hyper-lengthening in sir4Δ cells. This experiment was performed as described in Figure 2A, but the liquid culture passaging method described in Figure 3A was used instead of re-streaking single colonies on plates. (C) TLC1(Ku)3 does not cause telomere hyper-lengthening in sir2Δ or sir3Δ cells, and tlc1Δ48 causes greater telomere shortening in rif1Δ and rif2Δ cells than in wild-type cells. This experiment was performed as described in Figure 3B except that cells were passaged to ∼250 generations by re-streaking on plates rather than passaging in liquid cultures.
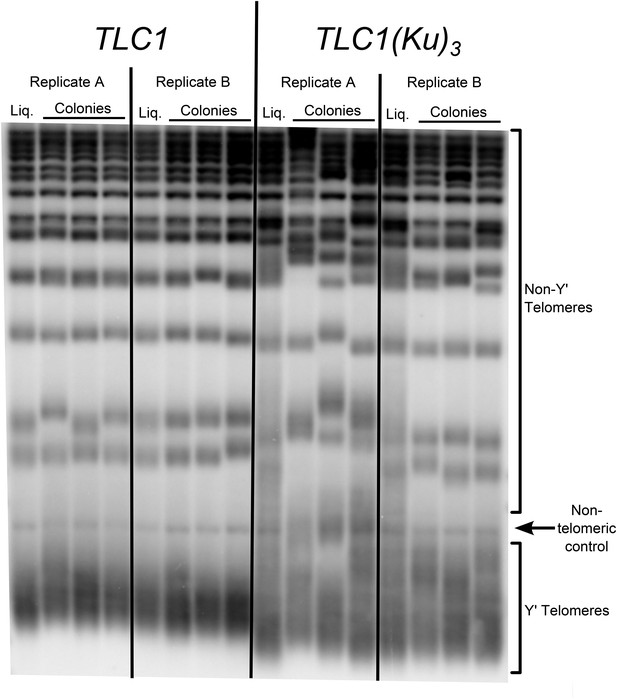
Three Ku-binding sites in yeast telomerase RNA increase telomere-length heterogeneity.
Cells were initially serially passaged in liquid culture as described in Figure 3A and then ∼25 generations before the end of passaging, liquid cultures were plated for single colonies. Genomic DNA was isolated from both the liquid-passaged cultures (‘Liq.’) and from cells cultured from the (clonal) colonies from solid medium (‘Colonies’).
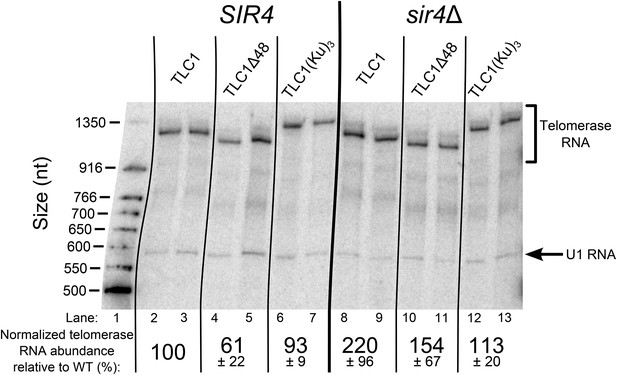
TLC1 RNA abundance is largely unaffected in TLC1(Ku)3 cells and is not decreased in sir4Δ cells.
Total RNA was isolated from the cells used in the experiment described in Figure 3B and subjected to Northern blot analysis. The pairs of lanes on the Northern blot represent two independent sets of biological replicates. The blot was probed for TLC1 and for the U1 snRNA. Telomerase RNA abundance was normalized to U1 and is expressed relative to the SIR4 TLC1 condition. The values shown are averages of these two replicates and another set biological replicates from a separate Northern blot, ± the standard deviation.
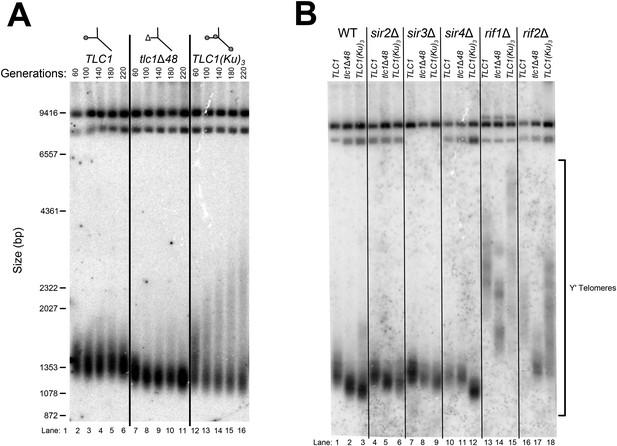
TLC1(Ku)3 causes Y′-telomere shortening and hyper-lengthening, while deletion of RIF1 or RIF2 causes Y′-telomere hyper-lengthening.
https://doi.org/10.7554/eLife.07750.009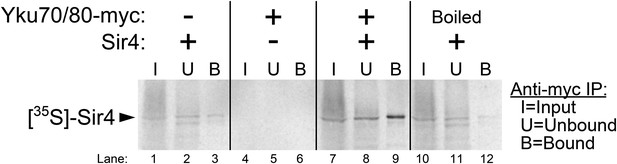
Purified Ku binds Sir4 in vitro.
35S-methionine-labeled Sir4 was synthesized in vitro in a rabbit reticulocyte lysate transcription/translation system (RRL) to which purified Ku heterodimer, bearing a 2myc epitope on the C-terminus of Yku80, was added. After Sir4 synthesis, the RRL was subjected to anti-myc immunoprecipitation. The input, unbound, and bound fractions were run on an SDS polyacrylamide gel, which was imaged by autoradiography.
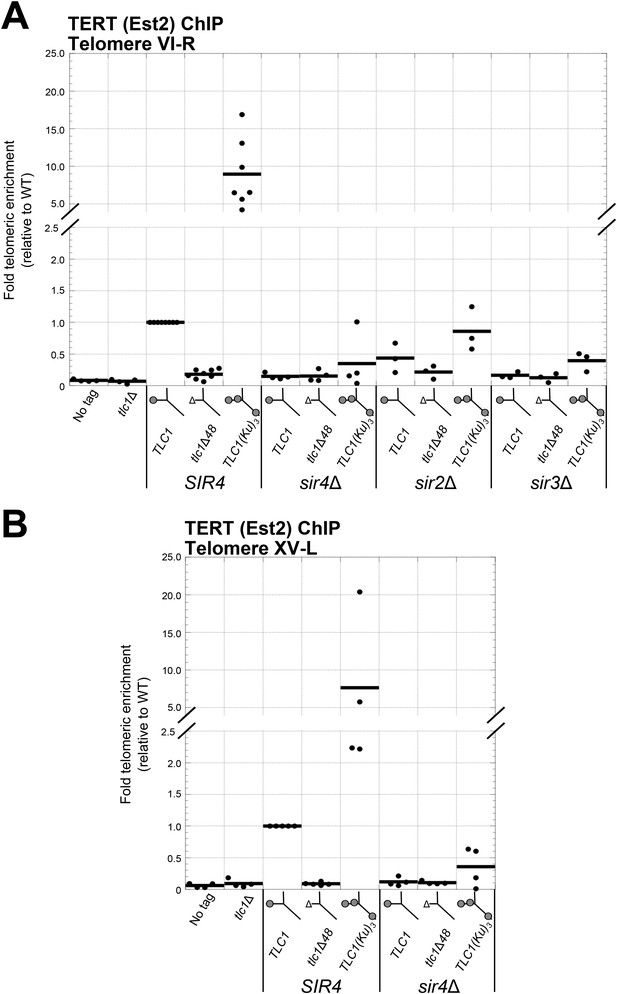
Ku-mediated telomerase recruitment to telomeres requires SIR4.
(A, B) In strains similar to those used in Figure 3B, TERT (Est2) was expressed from its endogenous locus bearing a C-terminal 13myc tag, separated by an 8-glycine linker. TLC1, TLC1Δ48, and TLC1(Ku)3 were expressed as in Figure 3B, but cells were not passaged after loss of the pTLC1-URA3 cover plasmid. Cells were crosslinked and subjected to chromatin immunoprecipitation (ChIP) using the myc epitopes on TERT, as described (Fisher et al., 2004). Telomeric enrichment was measured using quantitative real-time PCR (qPCR) amplicons close to telomere VI-R (A) and telomere XV-L (B). An amplicon at the ARO1 locus was used as a non-telomeric control locus. The thick horizontal lines on the graphs represent averages of three to five independent biological replicates, which themselves are indicated by black dots.
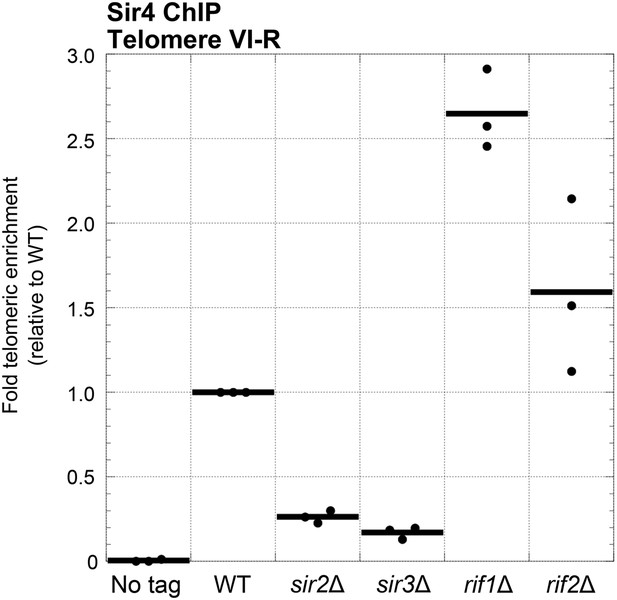
Sir4 binding to telomeres is decreased in sir2Δ and sir3Δ cells and increased in rif1Δ and rif2Δ cells.
Sir4 bearing a C-terminal 13myc tag on an 8-glycine linker was expressed from its endogenous chromosomal gene locus. Cells were crosslinked and subjected to ChIP using the myc epitopes. Telomere VI-R enrichment was measured using real-time quantitative PCR as in Figure 5A. The thick horizontal lines on the graph represent averages of three independent biological replicates indicated by black dots.
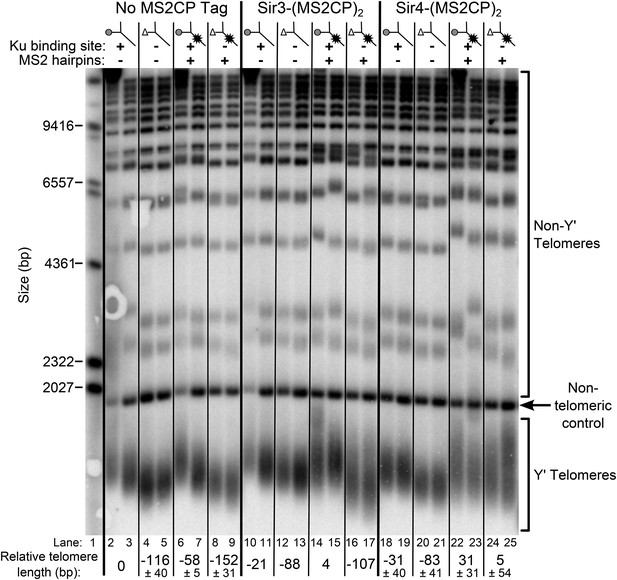
Tethering Sir4 to the tlc1Δ48 RNA restores telomeres to wild-type length.
Using the same tlc1Δ pTLC1-URA3 strain background from Figure 2A, Sir3 and Sir4 were expressed from their endogenous loci bearing C-terminal (MS2CP)2 tags, separated by an 8-glycine linker. These strains were transformed with CEN plasmids containing either TLC1, tlc1Δ48, TLC1(MS2)10, or tlc1Δ48(MS2)10. Cells were then cured of the pTLC1-URA3 cover plasmid and passaged as in Figure 2A. Each pair of lanes represents two independent biological replicates, and the relative telomere-length values are averages of the two replicates. In the no MS2 coat protein (MS2CP) tag and Sir4-(MS2CP)2 conditions, values from a third set of replicates were included in the average, allowing for standard deviation to be calculated.
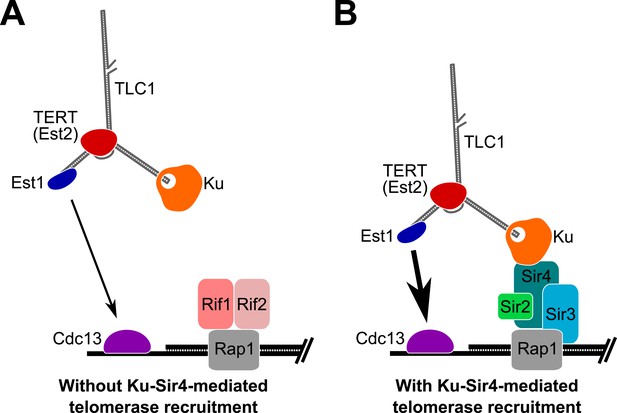
Model for Ku-Sir4 telomerase recruitment to telomeres and its role in telomere-length regulation in Saccharomyces cerevisiae.
Telomerase has previously been shown to extend a telomere infrequently and shorter telomeres are preferentially extendable. We propose here that Ku recruits telomerase to telomeres by binding Sir4. Since it has been shown that Rif1 and Rif2 compete with Sir4 and Sir3 for binding telomere-bound Rap1, the Ku-Sir4 telomerase recruitment pathway is inhibited by Rif1 and 2, providing a simple mechanistic explanation for one way in which Rif proteins function to inhibit telomerase action at telomeres. (A) Ku recruitment of telomerase via Sir4 is inhibited by Rif1/2 competition for Rap1 binding with Sir4. In situations where Ku-Sir4-mediated telomerase recruitment does not occur, Est1-Cdc13-mediated telomerase recruitment can still happen, although with low efficiently. (B) When telomerase is recruited to a telomere through the Ku-Sir4 pathway, subsequent Est1-Cdc13-mediated recruitment to the end of the telomere becomes more efficient, resulting in increased telomerase extension of telomeres. The counterbalancing of Ku-Sir4 telomerase recruitment and Rif1/Rif2 occlusion of Sir4 binding to Rap1 may represent a system for maintaining telomere-length homeostasis in yeast.
Tables
Average Yʹ telomere length in sirΔ cells containing TLC1, tlc1Δ48, or TLC1(Ku)3
| SIR Genotype | TLC1 Genotype | ||
|---|---|---|---|
| TLC1 | tlc1Δ48 | TLC1(Ku)3 | |
| SIR | 0 | −86 ± 23† | Dysregulated* |
| sir4Δ | −53 ± 13† | −71 ± 26† | −148 ± 36‡ |
| sir2Δ§ | −41 ± 16 | −50 ± 74 | −71 ± 26 |
| sir3Δ§ | −51 ± 20 | −84 ± 14 | −123 ± 2 |
-
The weighted-average mobility of the Yʹ telomeric restriction fragments was calculated as described in the ‘Materials and methods’. The numbers shown are averages of multiple biological-replicate samples ± standard deviation.
-
*
Yʹ telomere length was not quantified in this condition because signal from Yʹ telomere restriction fragments overlapped with that from the non-telomeric control fragment.
-
†
n = 6.
-
‡
n = 4.
-
§
n = 2.
Additional files
-
Supplementary file 1
(A) Yeast strains used in this study. (B) Plasmids used in this study.
- https://doi.org/10.7554/eLife.07750.015





