The β-hairpin of 40S exit channel protein Rps5/uS7 promotes efficient and accurate translation initiation in vivo
Figures
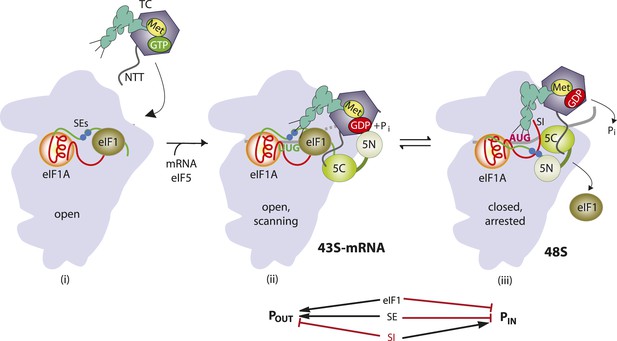
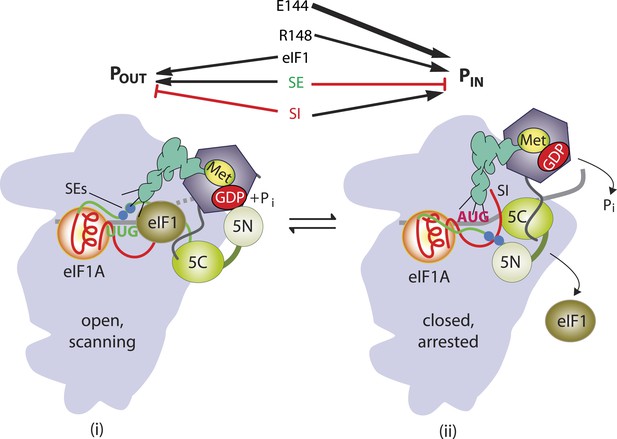
Model describing conformational rearrangements of the PIC during scanning and start codon recognition.
Assembly of the PIC, scanning and start codon selection in WT cells. (i) eIF1 and the scanning enhancer (SEs) elements in the CTT of eIF1A stabilize an open conformation of the 40S subunit to which the TC loads rapidly. (ii) The 43S PIC in the open conformation scans the mRNA for the start codon with Met-tRNAi bound in the POUT state. The GAP domain in the N-terminal domain of eIF5 (5N) stimulates GTP hydrolysis by the TC to produce GDP•Pi, but release of Pi is blocked. The unstructured NTT of eIF2β interacts with eIF1 to stabilize eIF1•40S association and the open conformation. (iii) On AUG recognition, Met-tRNAi moves from the POUT to PIN state, clashing with eIF1 and the CTT of eIF1A. Movement of eIF1 and the eIF1A CTT away from the P site disrupts eIF1's interaction with eIF2β-NTT, and the latter interacts with the eIF5-CTD. eIF1 dissociates from the 40S subunit, and the eIF5-NTD disengages from eIF2 and interacts with the eIF1A CTT instead, dependent on the SE elements, thereby facilitating Pi release from eIF2. The eIF5-CTD moves into the position on the 40S subunit previously occupied by eIF1 and blocks reassociation of eIF1. (Below) Arrows summarize that eIF1 and the eIF1A SE elements promote POUT and block transition to the PIN state, whereas the scanning inhibitor (SI) element in the NTT of eIF1A stabilizes the PIN state. (Adapted from Hinnebusch and Lorsch, 2012; Nanda et al., 2013).
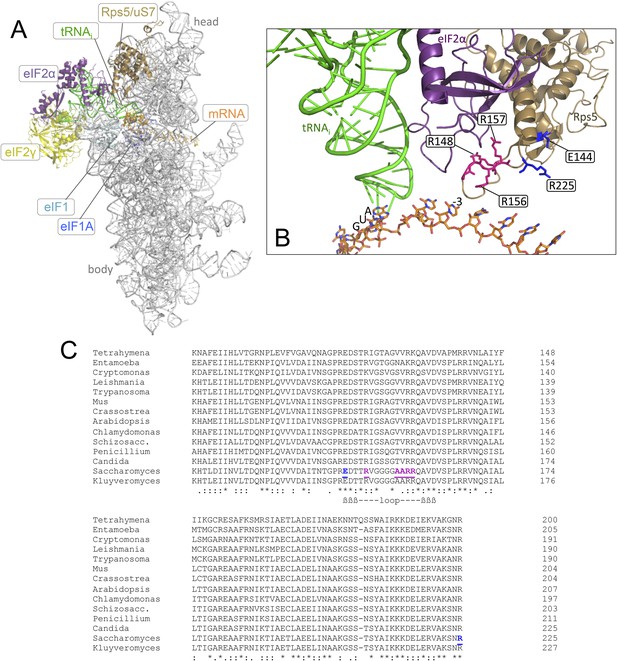
Location in the py48S PIC, and sequence conservation, of the Rps5 β-hairpin loop.
(A, B) Depiction of the partial yeast 48S PIC (PDB 3J81) showing Rps5 (gold), mRNA (orange), Met-tRNAi (green), eIF2α (purple), eIF2γ (yellow), eIF1 (cyan) and eIF1A (blue). For clarity other ribosomal proteins, eIF2β and putative eIF5 densities are not shown. Residues implicated here in AUG recognition are shown with stick side-chains and highlighted (as in panel C) in blue or pink. (C) Alignment of Rps5 sequences from diverse eukaryotes using the Clustal Omega algorithm (http://www.ebi.ac.uk/Tools/msa/clustalo/). The β- hairpin loop is annotated below the alignment and residues implicated in this study in AUG recognition are highlighted in blue or pink and underlined.
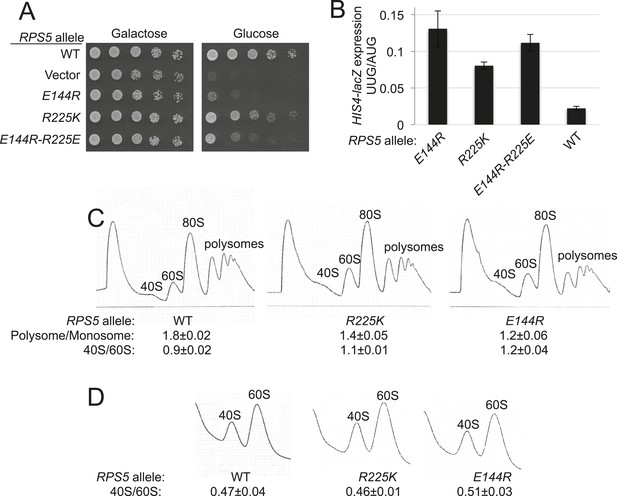
RPS5 mutations E144R and R225K impair translation initiation and elevate UUG initiation without reducing 40S subunit abundance in vivo.
(A) 10-fold serial dilutions of transformants of PGAL1-RPS5 his4-301 strain (JVY07) with the indicated plasmid-borne RPS5 alleles were spotted on synthetic medium supplemented with histidine and containing galactose (SGal + His + Ura + Trp) or glucose (SD + His + Ura + Trp) as carbon source and incubated at 30°C for 3 days (Glucose) or 4 days (Galactose). (B) Strains from (A) also harboring HIS4-lacZ reporters with AUG or UUG start codons (plasmids p367 and p391, respectively) were cultured in SD + His + Trp at 30°C to A600 of ∼1 and β-galactosidase specific activities were measured in WCEs in units of nanomoles of o-nitrophenyl- β-D-galactopyranoside (ONPG) cleaved per min per mg of total protein. Ratios of mean expression of the UUG and AUG reporters calculated from four transformants are plotted with error bars (indicating S.E.M.s). (C) Strains from (A) were cultured in SD + His + Ura + Trp at 30°C to A600 of ∼1, and cycloheximide was added prior to harvesting. WCEs were separated by sucrose density gradient centrifugation and scanned at 254 nm to yield the tracings shown. Mean Polysome/Monosome and 40S/60S ratios (and S.E.M.s) from four replicates are indicated. Student's t-test indicates that the mean values for polysome/monosome ratio in the RPS5 mutants are reduced significantly from the WT (p < 0.0005). (D) Similar to (C) but the cultures were not treated with cycloheximide and lysed in buffers without MgCl2 to allow separation of the dissociated ribosomal subunits.
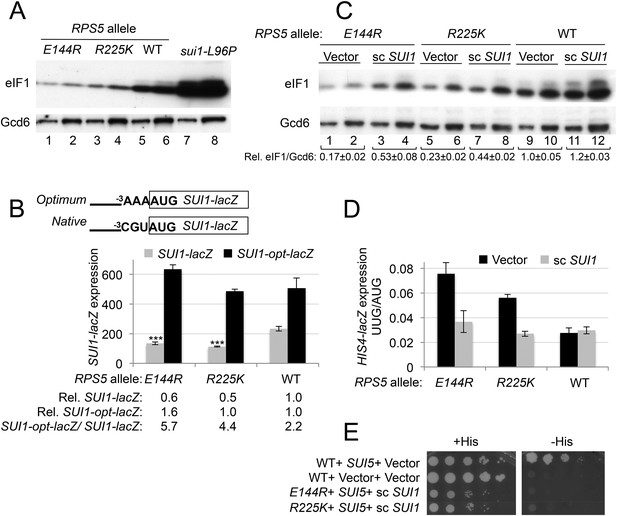
RPS5 mutations E144R and R225K exacerbate poor context at the native SUI1 AUG to reduce eIF1 expression and indirectly confer Sui- phenotypes, but evoke Ssu- phenotypes when eIF1 abundance is boosted.
(A) WCEs of strains from Figure 3A, and from sui1-L96P strain H4564, were subjected to Western analysis using antibodies against eIF1 or Gcd6 (as loading control). Two amounts of each extract differing by a factor of two were loaded in successive lanes. (B) Strains from Figure 3A also harboring SUI1-lacZ (pPMB24) or SUI1- opt-lacZ (pPMB25) reporters were cultured and assayed for β-galactosidase activities as described in Figure 3B. Mean expression levels and S.E.M.s from four transformants are plotted, and relative (Rel.) mean expression levels normalized to that of the WT strain are listed below the histogram. Student's t-test indicates that the mean values for SUI1-lacZ expression in the RPS5 mutants are reduced significantly from the WT (***p < 0.0005). (C) WCEs of strains from Figure 3A also harboring sc SUI1 plasmid pPMB21 or empty vector were subjected to Western analysis as in (A). Signal intensities were quantified and mean eIF1/Gcd6 ratios are listed below the blot with S.E.Ms (D) HIS4-lacZ reporters with AUG or UUG start codons were assayed in strains from (C) as in Figure 3B. (E) his4-301 strains with the indicated WT or mutant RPS5 alleles (from Figure 3A) harboring sc SUI1 plasmid pPMB21, SUI5 plasmid p4281, or empty vectors were spotted on SD plates containing (SD + His) or lacking histidine (SD-His) and incubated for 3 days and 5 days, respectively.
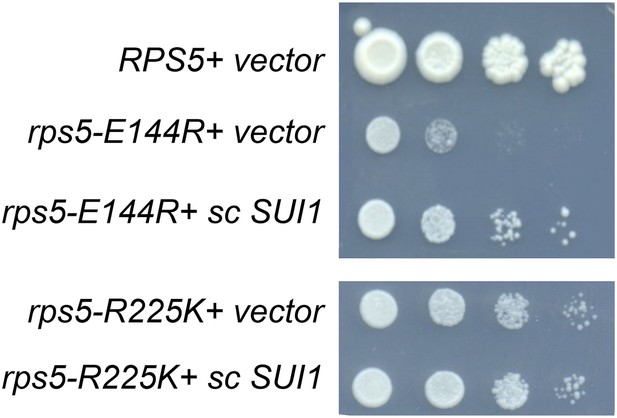
Increased SUI1 gene dosage partially rescues the Slg- phenotype of RPS5 mutations E144R and R225K.
10-fold serial dilutions of transformants of strains containing the indicated RPS5 alleles from Figure 3A harboring sc SUI1 plasmid pPMB21 or empty vector were spotted on SD + His + Ura and incubated at 30°C for 3 days. All strains were spotted in parallel on plates of identical medium.
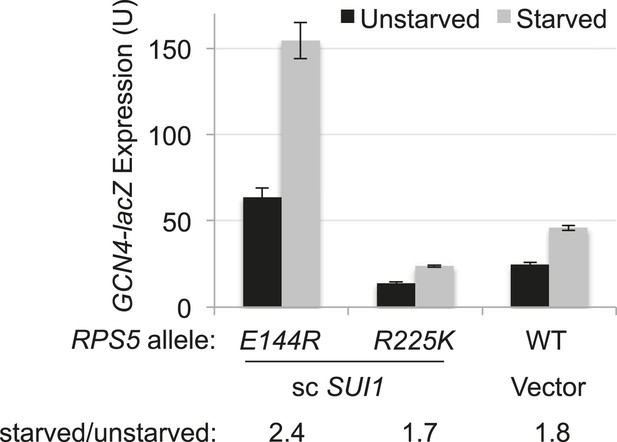
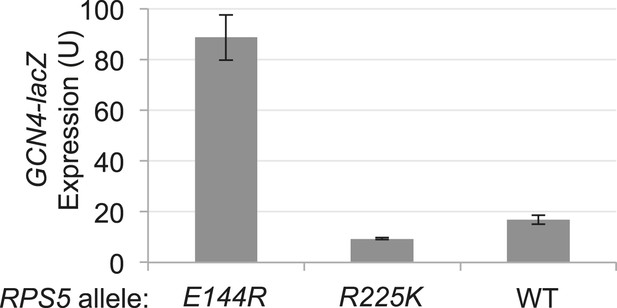
RPS5 mutation E144R confers a Gcd- phenotype, derepressing GCN4-lacZ expression with restored eIF1 expression.
Strains of the indicated RPS5 genotype from Figure 3A harboring either sc SUI1 plasmid pPMB21 or empty vector, and the WT GCN4-lacZ reporter on plasmid p180, were cultured in duplicate sets in SD + His at 30°C. One set (unstarved) was cultured to A600 of ∼1 and harvested. When the second set (starved) reached A600 of ∼0.5, sulfometuron methyl (SM) was added to 0.5 μg/ml and grown for 6 hr and harvested.β-galactosidase specific activities were measured in WCEs as described in Figure 3B. Mean expression levels and S.E.M.s were calculated from four transformants.
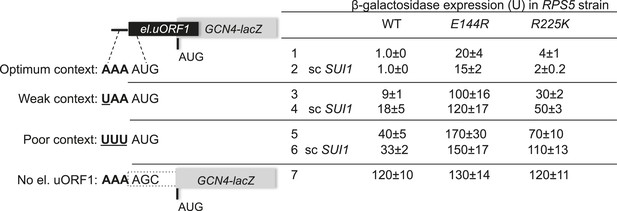
RPS5 mutations E144R and R225K confer strong leaky scanning of GCN4 uAUG-1 in vivo.
β-galactosidase activities were measured in WCEs of strains from Figure 4C harboring the scSUI1 plasmid (as indicated) and el.uORF1 GCN4-lacZ reporters pC3502, pC4466, or pC3503 containing, respectively, the depicted optimum, weak, or poor context of uAUG-1; or uORF-less GCN4-lacZ reporter pC3505 with a mutated uAUG-1. Mean expression values with S.E.M.s were determined from four transformants as described in Figure 3B.
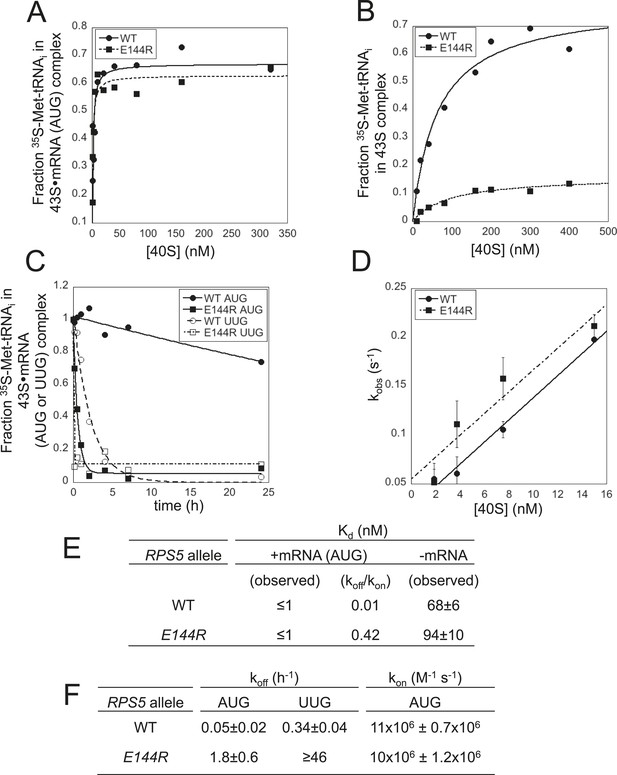
Rps5 Ssu- substitution E144R destabilizes the PIN state in vitro to a greater extent at UUG vs AUG start codons.
(A, B) Determination of Kd values for TC with [35S]-Met-tRNAi binding to 40S·eIF1·eIF1A complexes assembled with WT or E144R mutant 40S subunits and either mRNA (AUG) (A) or without mRNA (B). (C) Analysis of TC dissociation from 43S·mRNA complexes assembled with WT or E144R mutant 40S subunits and either mRNA(AUG) or mRNA(UUG). Representative curves selected from at least three independent experiments are shown. (D) Determination of kon values for TC binding to 40S·eIF1·eIF1A complexes from plots of observed rate constants (kobs) vs 40S concentration for WT or E144R mutant 40S subunits and mRNA(AUG). (E, F) Kd, koff and kon values with S.E.M.s determined in (A–D).
RPS5 mutation E144R confers a Gcd- phenotype, derepressing GCN4-lacZ expression.
Strains of the indicated RPS5 genotype from Figure 3A were transformed with p180 and analyzed for β-galactosidase expression as in Figure 3B. Mean expression levels and S.E.M.s were calculated from four transformants.
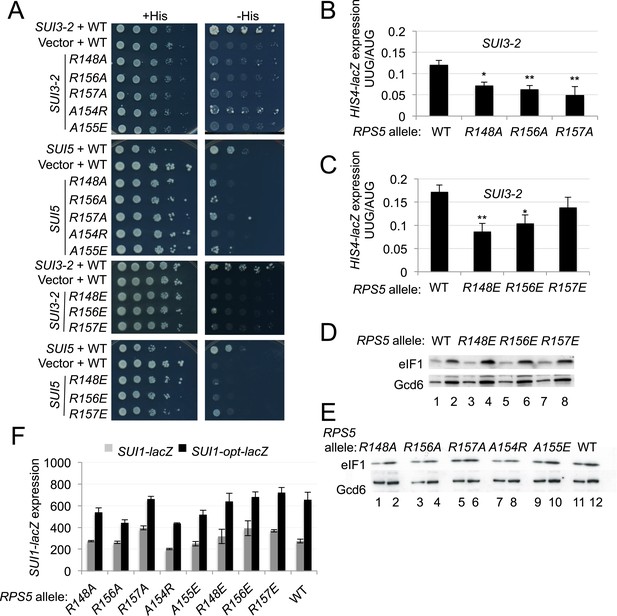
Substitutions in loop residues of the Rps5 β-hairpin confer Ssu- phenotypes.
(A) 10-fold serial dilutions of PGAL1-RPS5 his4-301 strain (JVY07) transformed with the indicated plasmid-borne RPS5 alleles and either SUI3-2 plasmid p4280, SUI5 plasmid p4281, or empty vector were spotted on SD + His + Ura (+His) or SD + Ura (−His) and incubated at 30°C for 3 days and 5 days, respectively. (B, C) Strains from (A) also harboring HIS4-lacZ reporters with AUG or UUG start codons (plasmids p367 and p391, respectively) were analyzed as in Figure 3B. Ratios of mean expression of the UUG and AUG reporters calculated from four transformants are plotted with S.E.M.s. Student's t-test indicates that the mean UUG/AUG expression in the RPS5 mutants is significantly reduced when compared to WT (*p < 0.05, **p < 0.005). (D, E) WCEs of his4-301 strains with the indicated RPS5 alleles were subjected to Western analysis as in Figure 4A. (F) WCEs of strains from (D, E) also harboring SUI1-lacZ (pPMB24) or SUI1-opt-lacZ (pPMB25) reporters were assayed for β-galactosidase activities as described in Figure 4B. Mean expression levels and S.E.M.s from four transformants are plotted.
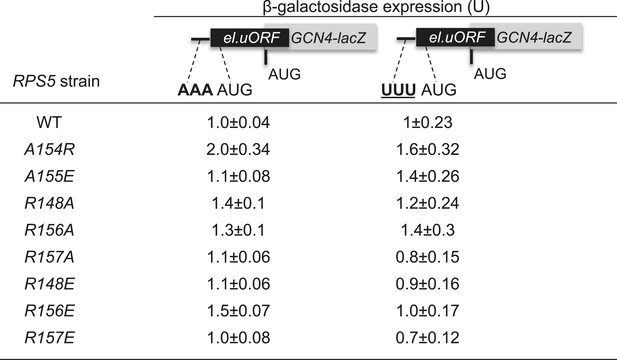
Substitutions in the loop of the Rps5 β-hairpin do not increase leaky scanning of GCN4 uAUG-1.
Strains isogenic to those in Figure 3A but containing the indicated RPS5 alleles were transformed with the el.uORF1 GCN4-lacZ reporters containing optimum (pC3502) or poor (pC3503) contexts of uAUG-1 and analyzed for β-galactosidase activities as in Figure 3B. Mean expression values were determined from four independent transformants and normalized to the value obtained for the WT strain, and the normalized means and S.E.M.s are reported.
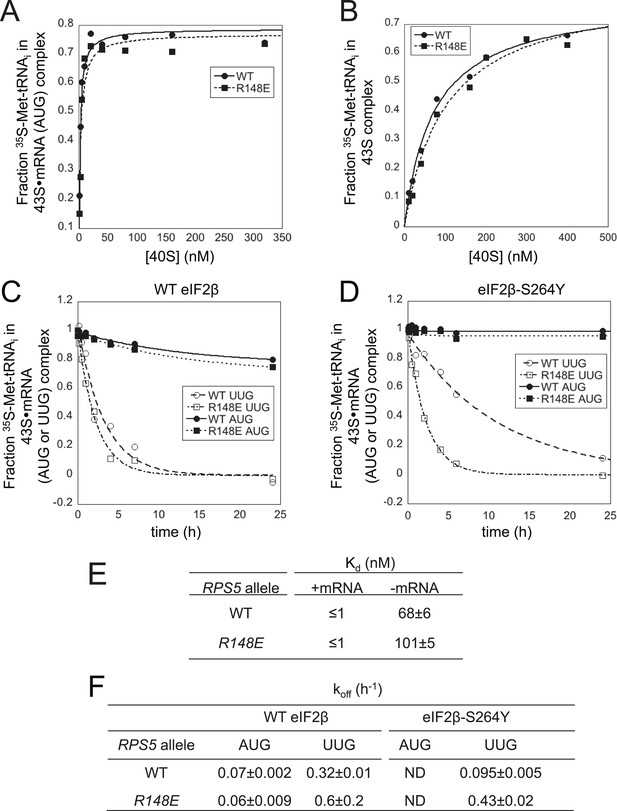
Rps5 Ssu- substitution R148E destabilizes PIN in vitro selectively at UUG codons.
(A, B) Determination of Kd values for TC with [35S]-Met-tRNAi binding to 40S·eIF1·eIF1A complexes assembled with WT or R148E mutant 40S subunits and either mRNA (AUG) (A) or without mRNA (B). (C, D) Analysis of TC dissociation from 43S·mRNA complexes assembled with WT or R148E mutant 40S subunits and mRNA(AUG) or mRNA(UUG), conducted using WT eIF2 (C) or eIF2β-S264Y mutant eIF2 (D). Representative curves selected from at least three independent experiments are shown. (E, F) Kd, koff values with S.E.M.s determined in (A–D). ND, no dissociation observed.
β-hairpin of Rps5 has a critical role in start codon recognition during translation initiation by stabilizing initiator tRNA binding to the pre-initiation complex.
Model summarizing the role of the conserved β-hairpin residues in Rps5 in start codon recognition. See Figure 1 for description of the open/POUT and closed/PIN states of the PIC and roles of eIF1 and the SE/SI elements of eIF1A in regulating conformational rearrangements and reactions accompanying AUG recognition. Results from this study indicate that Rps5 β-hairpin residues E144 and R148 function in stabilizing the PIN conformation of TC binding, with E144 having a stronger effect, as indicated by the thicker arrow.
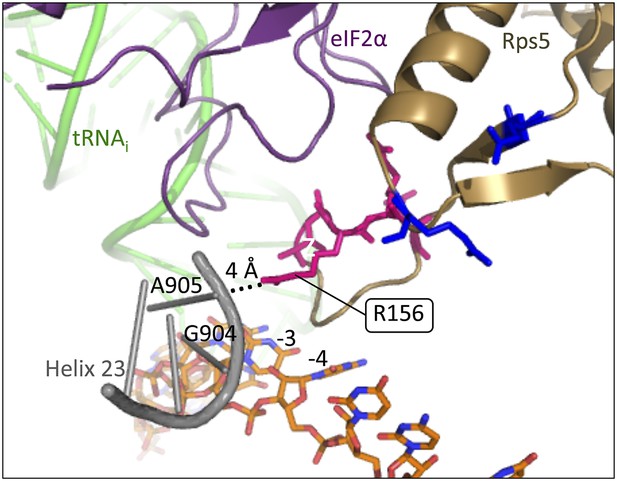
Rps5 β-hairpin loop is in proximity to rRNA helix 23.
Depiction of the py48S PIC (PDB 3J81) showing Rps5 in gold, mRNA in orange, Met-tRNAi in green, eIF2α in purple and rRNA residues of helix 23 in grey. The backbone of helix 23 is ∼4 Å from Rps5 loop residue R156 (indicated by dotted line), and rRNA residue G904 is within 3.6 Å of the −3 nucleotide in the mRNA.
Additional files
-
Supplementary file 1
Phenotypes of RPS5 mutants.
- https://doi.org/10.7554/eLife.07939.017
-
Supplementary file 2
Yeast strains employed in this study.
- https://doi.org/10.7554/eLife.07939.018
-
Supplementary file 3
Plasmids employed in this study.
- https://doi.org/10.7554/eLife.07939.019
-
Supplementary file 4
Oligonucleotide primers employed for mutagenesis in this study.
- https://doi.org/10.7554/eLife.07939.020





