The sterol-responsive RNF145 E3 ubiquitin ligase mediates the degradation of HMG-CoA reductase together with gp78 and Hrd1
Figures
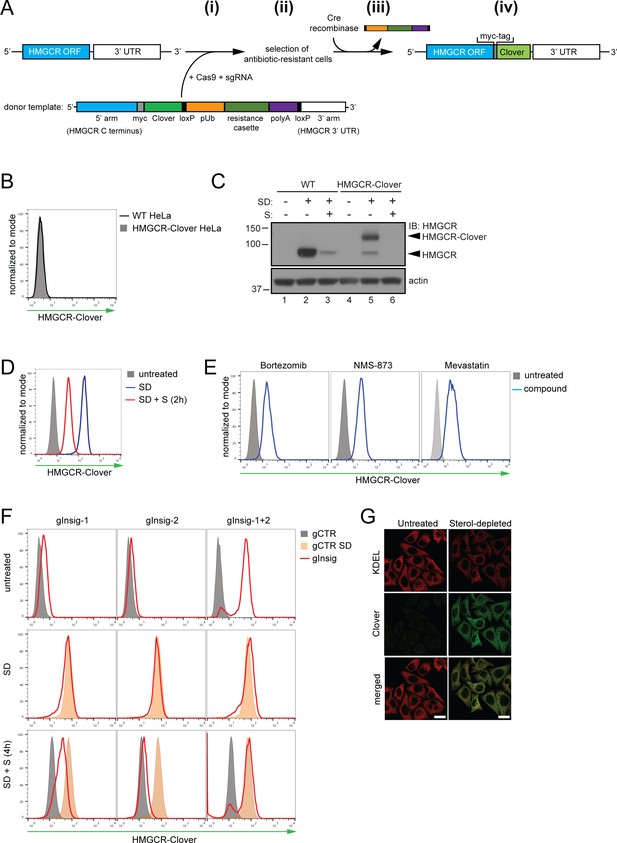
Fluorescent protein tagging of endogenous HMGCR generates a cholesterol-sensitive dynamic reporter.
(A) Schematic showing generation of the HMGCR-Clover reporter. (i) The endogenous HMGCR locus of HeLa cells was modified by transfection of Cas9, sgRNA and a donor template. The 5’ and 3’ arm of the donor template were designed as homologous sequences encoding the C-terminal region and 3’ UTR of the HMGCR gene. The C-terminal Clover tag (green) was appended in frame to the ORF of HMGCR (blue) including a myc-tag (grey) as spacer and an antibiotic resistance cassette flanked by loxP sites. (ii) Cells having stably integrated the recombination construct were enriched by antibiotic selection. (iii) The resistance cassette was removed by transient transfection of Cre recombinase to yield endogenous, C-terminally modified HMGCR (iv). pUb, ubiquitin promoter; ORF, open reading frame; UTR, untranslated region. (B – E) The HMGCR-Clover reporter phenocopies untagged HMGCR. (B) HMGCR-Clover expression (grey shaded histogram) as detected by flow cytometry under sterol-replete conditions. (C) Immunoblot of HMGCR in sterol-depleted (SD) HeLa WT vs. HMGCR-Clover cells -/+sterols (S) for 2 hr. For sterol depletion, cells were switched to SD medium (10% LPDS, mevastatin (10 μM), mevalonate (50 μM)) for 16 hr. Whole-cell lysates were separated by SDS-PAGE and HMGCR(-Clover) detected with an HMGCR-specific antibody. (D) Cytofluorometric analysis of HMGCR-Clover HeLa cells cultured in sterol-replete (shaded histogram) vs. sterol-depleted medium (SD) (16 hr, blue line histogram). Sterols (S) (2 µg/ml 25-hydroxycholesterol, 20 µg/ml cholesterol) were added back for 2 hr (red line histogram). (E) Flow cytometric analysis of HMGCR-Clover cells treated overnight with Bortezomib (25 nM), mevastatin (10 μM), or NMS-873 (10 μM) for 8 hr. (F) CRISPR/Cas9-mediated depletion of Insig-1 and −2 together (red line histogram) induce a dramatic increase in HMGCR-Clover expression, equivalent to sterol depletion (SD, orange shaded). HMGCR-Clover cells transiently expressing the indicated Insig-1/2 specific sgRNAs (four sgRNAs per gene) were treated as in (D) and, where indicated, sterols (S) added back for 4 hr (SD + S, bottom row). Representative of ≥3 independent experiments. (G) Immunofluorescence analysis of HMGCR-Clover and KDEL (ER marker) expression, showing co-localisation in sterol-depleted (SD, 16 hr) HMGCR-Clover HeLa cells. Scale bar = 20 μm.
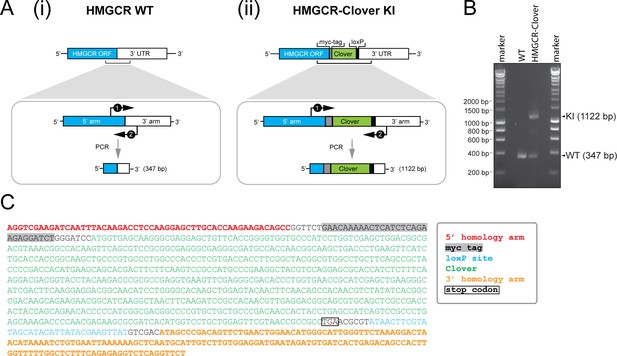
Genotyping of HMGCR-Clover knock-in cells.
(A) Schematic representation of PCR amplification of the genomic locus targeted for Clover insertion in HMGCR-Clover knock-in (KI) cells. Primers (‘1’, ‘2’) specifically annealing in the 5’ and 3’ homology arms flanking the Clover gene sequence were used to amplify a region encompassing 347 bp (i) in the WT allele and 1122 bp (ii) if the Clover insert is present. (B) The PCR reaction described in (A) was performed using genomic DNA derived from parental (WT) and HMGCR-Clover knock-in HeLa cells. Amplicons derived from the unmodified (WT, 347 bp) and HMGCR-Clover knock-in allele (KI, 1122 bp) are indicated. As described in Figure 1C, HeLa HMGCR-Clover KI cells retain at least one unmodified HMGCR allele in addition to endogenously Clover-tagged HMGCR. (C) Genetic profiling of the HMGCR-Clover KI locus in HMGCR-Clover HeLa cells. The longer 1122 bp PCR product, obtained as described in (A), was isolated by agarose gel electrophoresis and confirmed by Sanger sequencing.
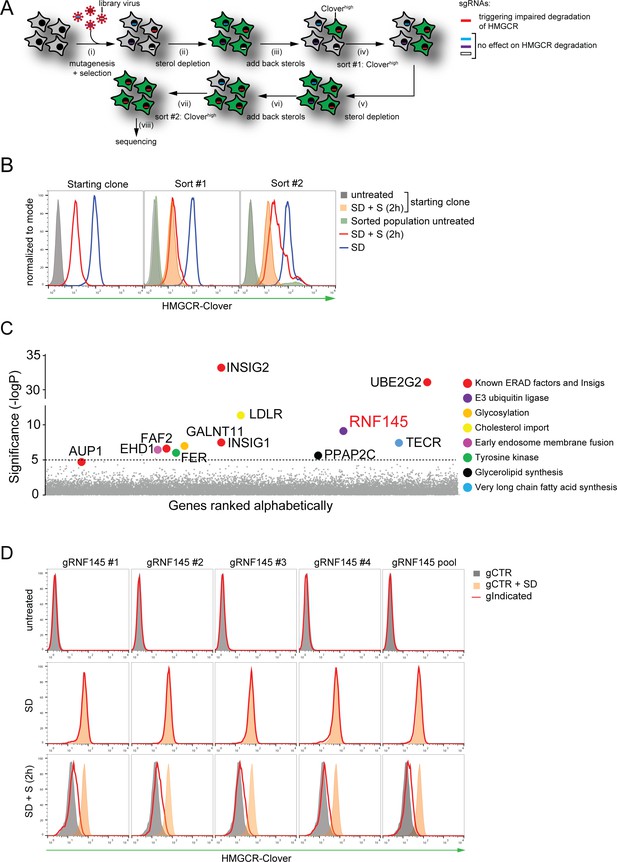
Genome-wide CRISPR knockout screen identifies a role for RNF145 in the sterol-dependent degradation of HMGCR.
(A - B) Schematic view of the CRISPR/Cas9 knockout screen workflow and FACS enrichment. (A) HMGCR-Clover HeLa cells transduced with a genome-wide sgRNA library targeting 19930 genes (i) were subjected to overnight sterol-starvation followed by sterol repletion for 5 hr (ii, iii, v, vi). Mutants unable to degrade HMGCR-Clover despite sterol repletion (Cloverhigh) were enriched by two sequential rounds of FACS (iv, vii) and candidate genes identified by deep sequencing (viii). (B) Enrichment of HMGCR-Clover mutants after sort #1 and sort #2 (red line histograms, corresponding to steps ‘iv’ and ‘vii’ in Figure 2A) as determined by flow cytometry. Cells were treated as described in Figure 1D. SD, sterol-depleted; S, sterols. (C) Candidate genes identified in the genome-wide knockout screen. Genes scoring above the significance threshold of - logP ≥5 (dotted line) and AUP1 (- logP = 4.7) are highlighted. (D) RNF145 depletion mildly impairs sterol-accelerated HMGCR-Clover degradation. HMGCR-Clover cells transiently expressing four independent RNF145-specific sgRNAs (gRNF145#1–4, red line histogram), individually or as a pool, vs. sgB2M (gCTR) were sterol-depleted overnight (middle row, SD) and re-examined by flow cytometry following 2 hr sterol addition (bottom row, SD + S). Representative of ≥3 independent experiments.
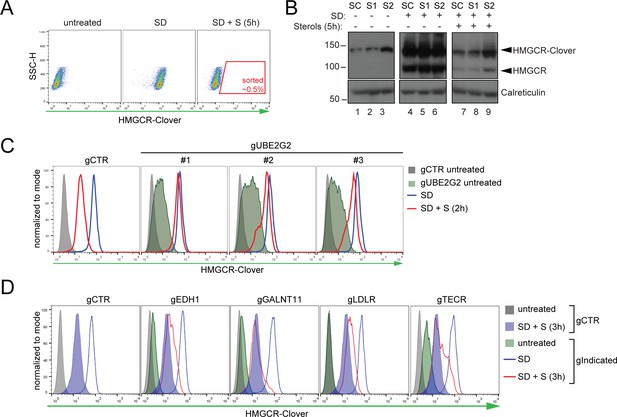
Genome-wide screen for proteins involved in HMGCR degradation.
(A) Gating strategy to enrich for Cloverhigh mutants with impaired sterol-induced HMGCR-Clover degradation. HeLa HMGCR-Clover cells mutagenized with a genome-wide sgRNA library were subjected to overnight sterol depletion (SD) before adding back sterols (SD + S) for 5 hr. Typically, the highest ~0.5% of Cloverhigh cells were selected for enrichment (indicated). SSC-H, side scatter. (B) Immunoblot analysis for HMGCR-Clover enrichment after sort #1 (S1) and sort #2 (S2) as compared to the starting clone (SC). Cells were sterol-depleted (SD) overnight ± sterols (5 hr). (C) Flow cytofluorometric analysis of HMGCR-Clover cells transiently expressing three independent sgRNAs targeting UBE2G2 (gUBE2G2 #1–3) versus sgB2M (gCTR) after overnight sterol-depletion ± sterols (2 hr). (D) HMGCR-Clover cells were transfected with a pool of four sgRNAs for each indicated gene or a sgRNA against B2M (gCTR) and treated as in (C).
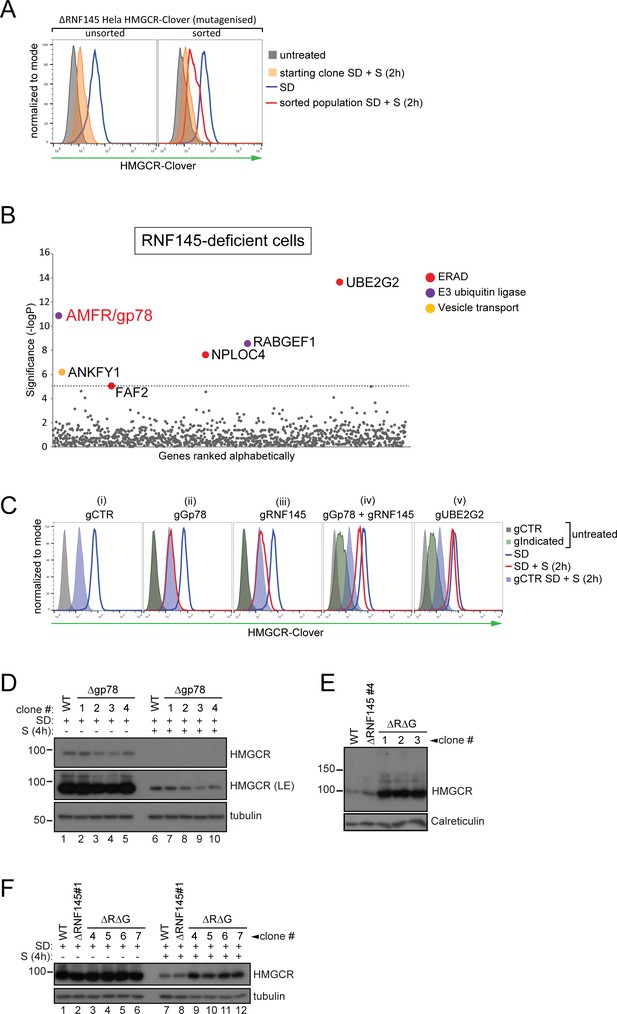
RNF145 together with gp78 are required for HMGCR degradation.
(A - B) FACS enrichment and scatter plot of candidate genes identified in the ubiquitome-targeted knockout screen. (A) HMGCR-Clover ΔRNF145#5 HeLa cells were mutagenized using a targeted ubiquitome-specific sgRNA library and mutant cells showing impaired sterol-dependent degradation of HMGCR-Clover were enriched by FACS. Enrichment is represented by a broad population of Cloverhigh cells in the presence of sterols (S, 2 hr) after overnight sterol depletion (SD, blue to red histogram). (B) Genes scoring above the significance threshold of - logP ≥5 (dotted line) are highlighted. (C - F) sgRNA targeting of gp78 together with RNF145 increases steady-state HMGCR-Clover and inhibits sterol-accelerated degradation of HMGCR-Clover in sterol-starved cells. (C) HMGCR-Clover cells transiently transfected with indicated sgRNAs were sterol-depleted (SD) overnight (blue line histogram) and sterols (2 µg/ml 25-hydroxycholesterol, 20 µg/ml cholesterol) added back (SD + S) for 2 hr (red line histogram or blue shaded histogram for sgB2M (gCTR)). Representative of ≥3 independent experiments. (D) Four independent gp78 knockout clones (#1–4) or WT cells were sterol-depleted (16 hr) ± S (4 hr) and HMGCR levels monitored by immunoblotting. LE, long exposure. (E) HMGCR steady-state levels in three RNF145/gp78 double knockout clones (ΔRΔG #1–3). (F) Four RNF145/gp78 double-knockout clones (ΔRΔG #4–7), RNF145 knockout, and WT cells were sterol-depleted (SD) overnight and HMGCR expression assessed ± sterols (S, 4 hr) by immunoblot analysis.
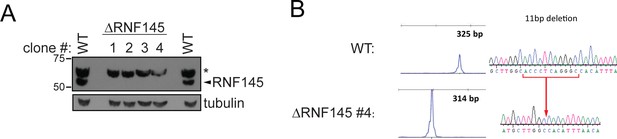
Validation of RNF145 knockout clones.
(A) Immunoblot for RNF145 in HeLa WT vs. four RNF145 knockout (ΔRNF145) clones, generated with two independent sgRNAs. Non-specific bands are indicated (*). (B) Genetic validation of RNF145 knockout clone #4 (ΔRNF145 #4). The genomic region surrounding the predicted sgRNA annealing site was amplified using fluorescent primers and amplicon size determined by capillary electrophoresis (Agilent Bioanalyzer 2100). Fluorescent traces are shown alongside amplicon sequences as obtained by Sanger sequencing, confirming an 11 base pair deletion (indicated).
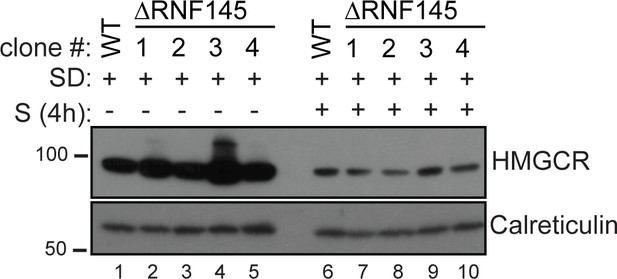
RNF145 loss is insufficient to block sterol-induced HMGCR degradation.
Cells were sterol-depleted (SD) overnight before addition of sterols (S, 4 hr). Whole-cell lysates from WT and four RNF145 knockout clones (#1–4) were separated by SDS-PAGE and HMGCR levels visualised by immunoblot analysis. Calreticulin serves as a loading control.
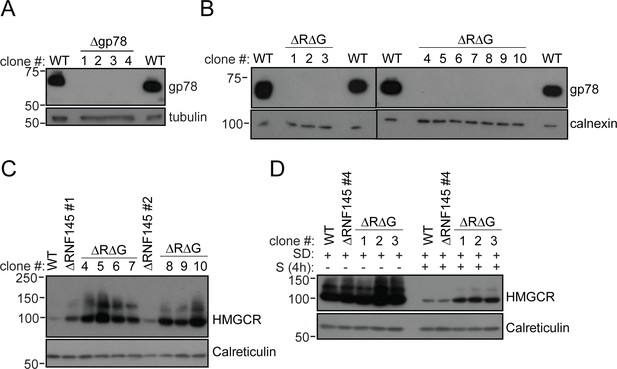
RNF145/gp78 double-knockout cells show increased HMGCR at steady-state and impaired sterol-induced HMGCR degradation.
(A) Immunoblot of four HeLa gp78 knockout clones derived by transfection with two independent gp78 sgRNAs. (B) Confirmation of gp78 loss in RNF145/gp78 knockout HeLa cells (ΔRΔG) derived from ∆RNF145 clones #1, #2, and #4 (for validation of RNF145 knockout see Figure 3—figure supplement 1). Calnexin serves as a loading control. (C) Steady-state expression of HMGCR in HeLa WT, ΔRNF145, and RNF145/gp78 double knockout clones (ΔRΔG) was determined by immunoblotting. (D) Indicated cell lines were sterol-depleted (SD) overnight ± sterols (S, 4 hr) and HMGCR detected by immunoblotting. Calreticulin serves as a loading control.
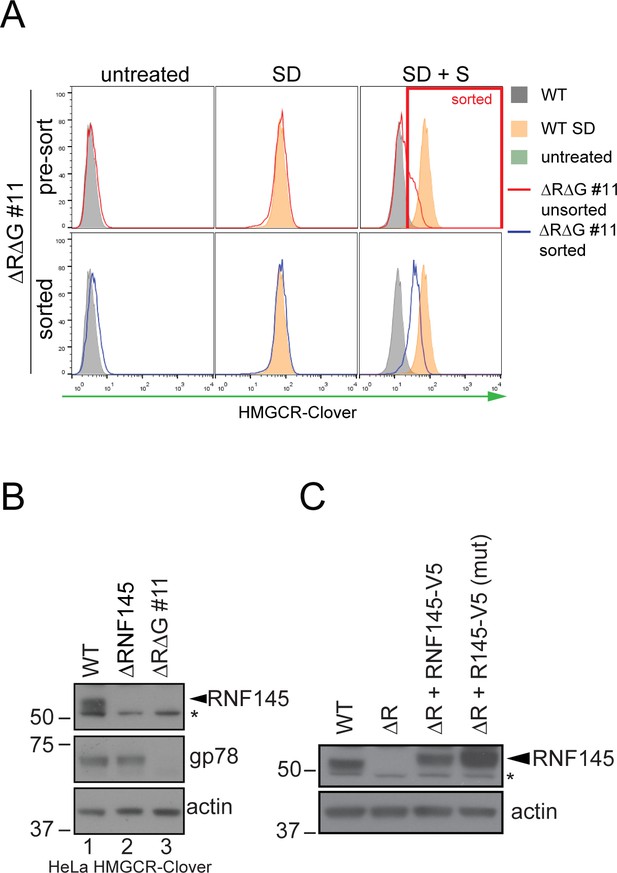
Establishment of RNF145/gp78 knockout HeLa HMCR-Clover and RNF145 complementation cell lines.
(A) HeLa HMGCR-Clover cells were transfected with RNF145 sgRNA#8 and gp78 sgRNA#4 (ΔRΔG #11, for sgRNAs used see Supplementary file 4 and 5) and knockout pools were enriched with puromycin. Eight days post transfection, cells were sterol-depleted (SD) overnight ± sterols (S, 2 hr). HMGCR-Cloverhigh cells were enriched by FACS (indicated). (B) RNF145 and gp78 levels in WT versus ∆RNF145 (clone #5) and RNF145/gp78-depleted (∆R∆G #11) HeLa HMGCR-Clover cells. (C) Stable genetic complementation of ΔRNF145 HeLa cells (ΔR, clone #4) by transduction with constructs encoding either RNF145-V5 (ΔR + RNF145-V5) or RNF145(C552A, H554A)-V5 (ΔR + RNF145-V5 (mut)). RNF145 variants were detected using an RNF145-specific antibody. Non-specific bands are indicated (*).
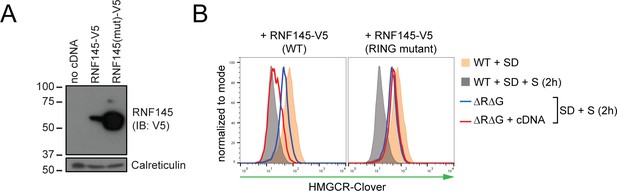
RNF145 E3 ligase activity is required for HMGCR degradation.
(A) Exogenous expression of RNF145 and RING-mutant RNF145 in HMGCR-Clover HeLa cells. RNF145/gp78 double-knockout HMGCR-Clover cells were transduced with lentivirus expressing either RNF145-V5 (WT) or a catalytically inactive RING domain mutant RNF145(C552A, H554A)-V5 cDNA (RING mutant) and cell lysates separated by SDS-PAGE and visualised by immunoblot analysis. IB, immunoblot. (B) Wild type, but not RING mutant RNF145, complements the RNF145-deficiency phenotype. RNF145/gp78 double-knockout HMGCR-Clover cells (ΔRΔG #11) were transduced with lentivirus expressing either RNF145-V5 or a catalytically inactive RING domain mutant RNF145(C552A, H554A)-V5 cDNA. Cells were sterol-depleted (16 hr) and after sterol repletion (2 hr), HMGCR-Clover levels were assessed by flow cytometry.
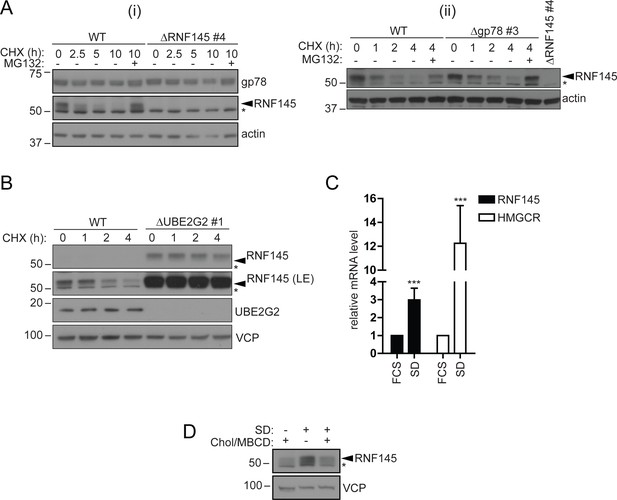
RNF145 is an intrinsically unstable, sterol-responsive E3 ligase.
(A and B) RNF145 has a short half-life and is auto-regulated by UBE2G2. (A) Translational shutoff analysis of gp78 in WT versus ΔRNF145 #4 (i) or RNF145 in Δgp78 #3 cells (ii) treated with cycloheximide (CHX, 1 µg/ml) ± MG132 (20 µg/ml) for the indicated times. Non-specific bands are indicated by an asterisk (*). Representative of ≥2 independent experiments. (B) Immunoblot analysis of WT and ΔUBE2G2 HeLa cells treated with CHX (1 µg/ml) for the indicated times. VCP serves as a loading control. LE, long exposure. An asterisk (*) signifies non-specific bands. (C and D) Sterol depletion induces transcriptional activation and increased levels of RNF145 protein. (C) Relative RNF145 and HMGCR mRNA levels as measured by quantitative PCR in HeLa cells grown in 10% FCS (FCS) or sterol-depleted (SD, 10% LPDS + 10 µM mevastatin + 50 µM mevalonate) for 48 hr. Mean ± S.D. (n = 4) and significance are shown, unpaired Students t-test: ***p≤0.001. (D) HeLa cells were grown under sterol-rich or -deplete conditions (±SD, as indicated) for 48 hr in the presence of mevastatin (10 µM) and mevalonate (50 µM) ± complexed cholesterol (Chol:MBCD, 37.5 µM). Whole-cell lysates were separated by SDS-PAGE and underwent immunoblot analysis. Non-specific bands are indicated (*). Representative of ≥2 independent experiments.
-
Figure 5—source data 1
Raw data from qPCR experiment in Figure 5C.
- https://doi.org/10.7554/eLife.40009.019
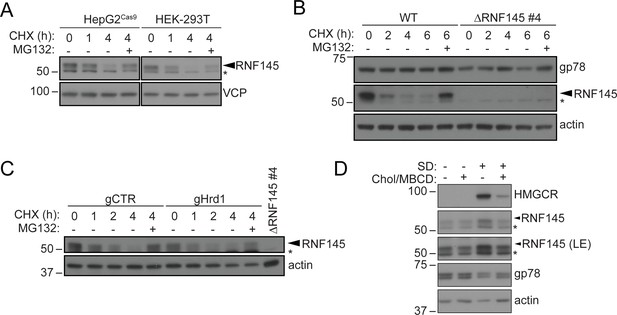
RNF145 is rapidly degraded by the ubiquitin proteasome system.
(A) Translation shutoff assay in HepG2Cas9 and HEK-293T cells. Cells were treated with cycloheximide (CHX, 1 µM) ± MG132 (20 µg/ml) for the indicated times and endogenous RNF145 levels determined by immunoblotting. (B) Gp78 is stable in the absence of RNF145. WT and ∆RNF145 #4 HeLa cells were cultured in the presence of CHX, (1 µM) ± MG132 (10 µg/ml) for 0–6 hr and gp78/RNF145 levels monitored by Western blotting. The asterisk (*) indicates a non-specific band. (C) HeLaCas9+ HMGCR-Clover were transiently transfected with a pool of four Hrd1-specific sgRNAs (gHrd1) or a B2M targeting control sgRNA (gCTR) and sgRNA containing cells enriched by puromycin selection. Cells were treated as in (B) for the indicated times. (D) HeLa cells were grown under sterol-rich or sterol-deplete conditions (±SD) for 48 hr ± complexed cholesterol (Chol/MBCD, 25 µM). SDS-PAGE and immunoblot analysis was performed on whole-cell lysates. Non-specific bands are indicated (*). LE, long exposure.
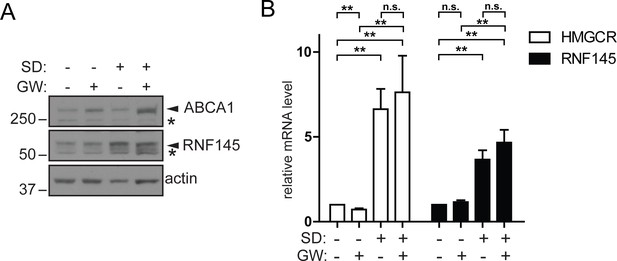
Increased RNF145 transcription upon sterol depletion is LXR-independent.
(A) Hela cells were sterol-depleted (SD, 10% LPDS + 10 µM mevastatin + 50 µM mevalonate) for 48 hr in the presence of the synthetic LXR ligand GW3965 (GW, 1 µM) or DMSO (vehicle control). Whole–cell lysates were analysed by SDS-PAGE and immunoblot assay. Representative of ≥3 independent experiments. (B) HeLa Cells were treated as described in (A) and the expression of indicated genes determined by quantitative PCR. All values were normalised to the steady-state, DMSO treated condition (bars 1 and 5). Mean ± S.D. (n = 3) and significance are indicated, unpaired Students t-test: **p≤0.01, ***p≤0.001, n.s. not significant.
-
Figure 5—figure supplement 2—source data 1
Raw data from qPCR experiment in Figure 5—figure supplement 2B.
- https://doi.org/10.7554/eLife.40009.018
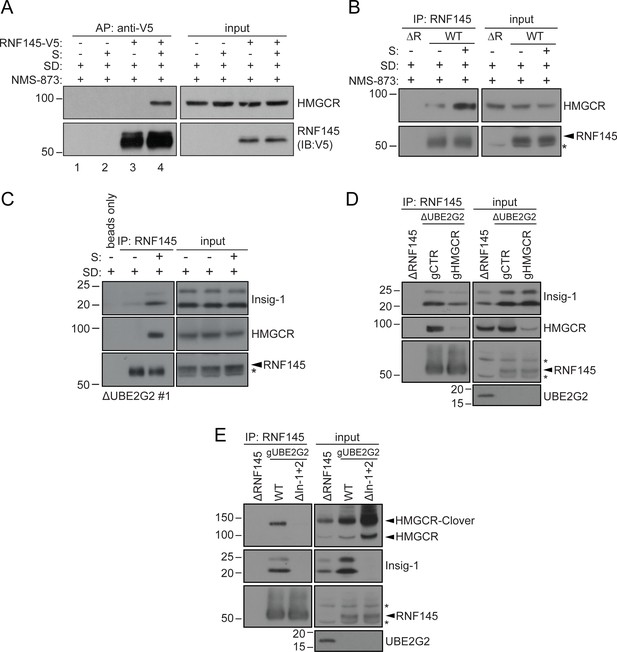
Endogenous RNF145 shows sterol-sensitive binding to Insig-1 and HMGCR.
(A) Exogenous RNF145 shows sterol-sensitive binding to HMGCR. RNF145 knockout cells stably reconstituted with RNF145-V5 (ΔR145 #4 + R145-V5, as shown in Figure 3—figure supplement 4C, lane 3) were sterol-depleted (SD, 20 hr) and, where indicated, sterols (S) added back for 1 hr in the presence of NMS-873 (10 µM, 1.5 hr). RNF145-V5 was affinity-purified (AP) and HMGCR detected by immunoblotting. Representative of ≥3 independent experiments. (B - C) Endogenous RNF145 shows sterol-sensitive binding to HMGCR and Insig-1. (B) HeLa WT or ΔRNF145 #4 (∆R) cells were treated as in (A), endogenous RNF145 was immunoprecipitated (IP), and RNF145 and HMGCR detected by immunoblot analysis. Non-specific bands are designated by an asterisk (*). Representative of ≥3 independent experiments. (C) HeLa UBE2G2 knockout cells (ΔUBE2G2 #1) were sterol-depleted (SD, 20 hr) and, where indicated, sterols (S) added for 1 hr. Endogenous RNF145 was affinity-purified and following SDS-PAGE separation, Insig-1 and HMGCR detected by immunoblot analysis. Representative of ≥2 independent experiments. (D - E) Insigs mediate binding between RNF145 and HMGCR. (D) HeLa UBE2G2 knockout cells (ΔUBE2G2 #1) transfected with a pool of four sgRNAs targeting HMGCR (gHMGCR) or sgRNA targeting B2M (gCTR) were enriched by puromycin selection and sterol-depleted for 20 hr, before adding back sterols for 1 hr + NMS-873 (10 µM, 1.5 hr). Endogenous RNF145 was immunoprecipitated and Insig-1 and HMGCR detected by immunoblot analysis. Non-specific bands are designated by an asterisk (*). (E) HMGCR-Clover HeLa WT or Insig-1+2 knockout (ΔIn-1+2) cells were transfected with a pool of three sgRNAs targeting UBE2G2 (gUBE2G2) and treated as in (D). Non-specific bands are designated by an asterisk (*).
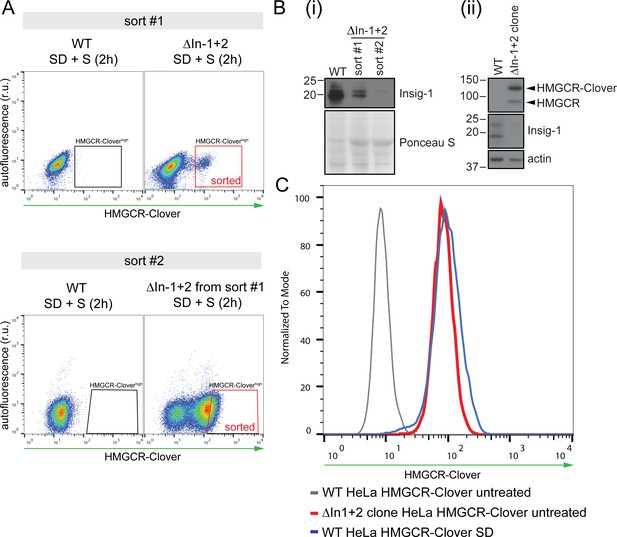
Generation of Insig knockout cell lines.
(A) HeLa HMGCR-Clover cells expressing Cas9 were transfected with four sgRNAs targeting Insig-1 and Insig-2 (ΔInsig-1+2), and selected with puromycin. Due to the low initial enrichment of Insig-1+2 double knockouts in this experiment, double knockouts were further enriched by two consecutive rounds of FACS (‘sort #1’ and ‘sort #2’), selecting cells with high constitutive HMGCR-Clover expression (‘sorted’, red box) after overnight sterol depletion (SD) and sterol repletion (S) for 2 hr. r.u., relative units. (B) Immunoblotting for endogenous Insig-1 in cells isolated from sort #1 and #2 in (A) (i) and in the Insig-1+2 double knockout clone (ΔIn-1+2 clone) isolated from the enriched population in sort #2 (ii). Insig-2 expression could not be assessed by immunoblotting due to the lack of a working Insig-2 antibody and/or low expression levels in these cells. (C) Validation of the Insig-1+2 double knockout clone isolated by single-cell sorting from the sort #2 population in (A). Loss of both Insigs was confirmed by presence of sterol-independent, constitutive expression of HMGCR-Clover due to de-repression of the SCAP-SREBP2 complex (red line). HMGCR-Clover expression under steady-state conditions in Insig-1+2 double knockout cells is comparable to that of the sterol-depleted (SD, 20 hr) parental WT cell line (blue line). Loss of Insig-1 in the Insig-1/2 double knockout single cell clone was further confirmed by immunoblotting (see (B) and Figure 6E).
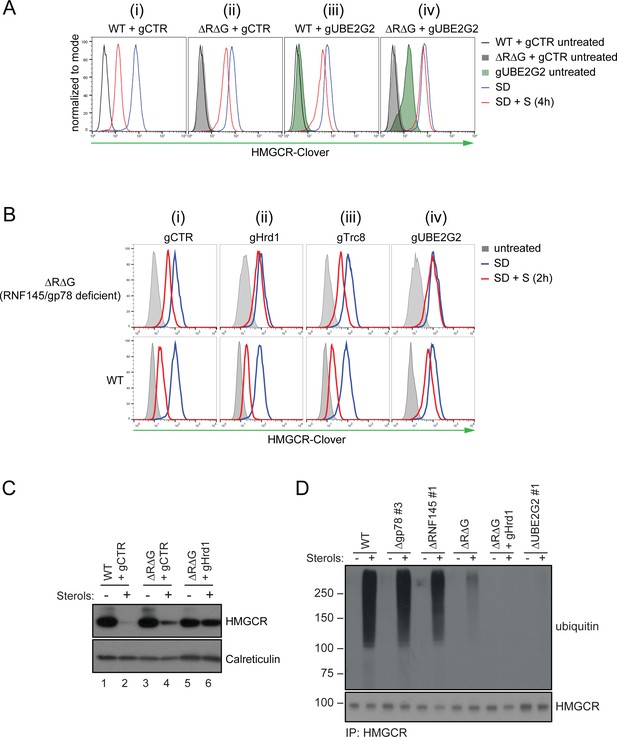
In the absence of RNF145 and gp78, Hrd1 targets HMGCR for ubiquitination and degradation.
(A) Loss of gp78, RNF145 and UBE2G2 exert an additive effect on HMGCR degradation. WT or RNF145/gp78 double knockout (ΔRΔG #11) HMGCR-Clover HeLa cells transiently expressing sgRNAs targeting UBE2G2 (gUBE2G2) or B2M (gCTR) were enriched by puromycin selection, sterol-depleted (SD) overnight and HMGCR-Clover expression assessed ± sterols (S, 4 hr). Representative of 3 independent experiments. (B and C) A targeted gene approach shows that loss of Hrd1 from RNF145/gp78 double knockout cells blocks sterol-accelerated degradation of HMGCR. (B) WT and ΔRNF145 Δgp78 (ΔRΔG #11) HMGCR-Clover cells transfected with sgB2M (gCTR), a pool of 3-4 sgRNAs targeting either Hrd1 (gHrd1), TRC8 (gTRC8) or UBE2G2 (gUBE2G2), were sterol-depleted (SD, 20 hr) and HMGCR-Clover expression assessed by FACS analysis ± sterols (S, 2 hr). (C) WT and RNF145/gp78 double knockout cells (ΔRΔG #7) were transfected with a pool of four Hrd1-specific sgRNAs or gCTR, sterol-depleted overnight before addition of sterols (4 hr) and analysis by SDS-PAGE and immunoblotting. RNF145 and gp78 knockout validation is shown in Figure 3—figure supplement 1B (ΔRNF145 #1) and Figure 3—figure supplement 3B (ΔRΔG #7), respectively. (D) RNF145, gp78 and Hrd1 are required for sterol-accelerated HMGCR ubiquitination. HMGCR was immunoprecipitated (IP) from the indicated cell lines grown in sterol-depleted media (20 hr) ± sterols (1 hr). MG-132 (50 µM) was added 30 min before sterol supplementation. Ubiquitinated HMGCR was detected using an anti-ubiquitin antibody.
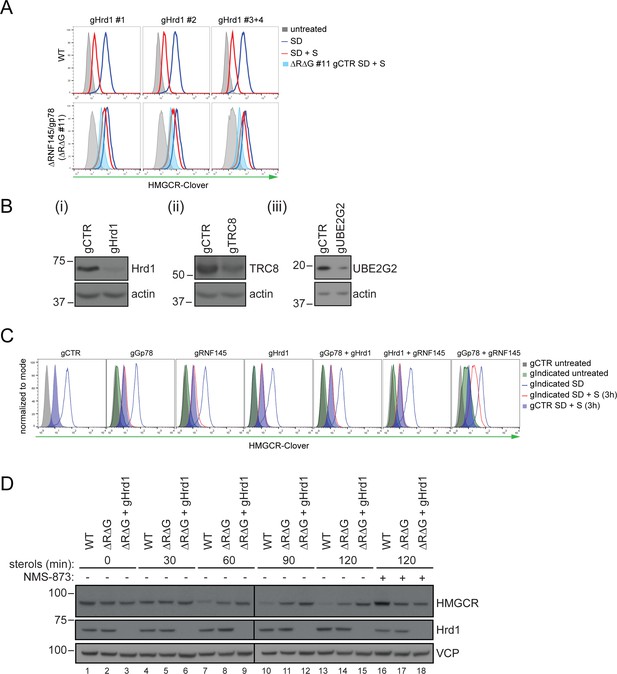
Combinatorial depletion of E3 ubiquitin ligases.
(A) WT or RNF145/gp78 knockout (∆R∆G #11) HeLa HMGCR-Clover cells transiently transfected with B2M-specific (gCTR) or Hrd1-specific (gHrd1 #1–4) sgRNAs were sterol-starved (SD, 20 hr) ± sterols (S, 2 hr). HMGCR-Clover expression was detected by FACS analysis. Cells transfected with gCTR are from the same experiment shown in Figure 7B and histograms (gCTR SD +S) were therefore re-plotted. (B) Validation of Hrd1 (i), TRC8 (ii), and UBE2G2 (iii) depletion in ∆R∆G #11 cells. Cell lines used in Figure 7B and Figure 7—figure supplement 1A were collected at steady-state and indicated proteins detected from whole-cell lysate by immunoblotting. A sgRNA targeting B2M (gCTR) served as a control. (C) WT HMGCR-Clover cells transfected with indicated sgRNA pools were sterol-depleted (SD) overnight ± sterols (S, 3 hr). Reporter expression was measured by FACS. (D) WT and ∆R∆G #7 HeLa cells transfected with gCTR (designated ∆R∆G) or a pool of four Hrd1-specific sgRNAs (∆R∆G + gHrd1) were sterol-depleted (20 hr) before addition of sterols for the indicated times ± NMS-873 (10 µM). Validation of RNF145 (∆RNF145 #1) and gp78 knockout clones (∆R∆G #7) can be found in Figure 3—figure supplements 1A and 3B, respectively.
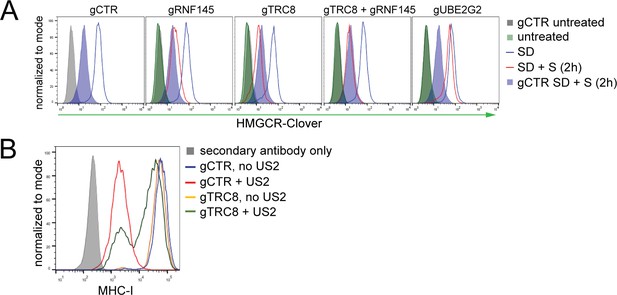
TRC8 depletion does not affect HMGCR-Clover degradation.
(A) Overnight sterol depletion (SD) ± sterols (S, 2 hr) in HeLa HMGCR-Clover cells transiently transfected with pools of indicated sgRNAs as described in 'Materials and methods'. (B) TRC8 knockdown was confirmed by US2-mediated TRC8-dependent downregulation of MHC-I. HeLa cells transiently expressing either B2M sgRNA (gCTR) or TRC8-sepcific sgRNAs (gTRC8) were selected for puromycin resistance and transduced with a lentiviral US2 and/or TRC8 construct 5 days post transfection. Cell-surface MHC-I staining and FACS analysis were performed on day 10 post transfection. US2 is a herpes cytomegalovirus (HCMV)-encoded gene product which directs TRC8 to degrade MHC-I in the ER lumen (Stagg et al., 2009; Hsu et al., 2015). Loss of TRC8 renders cells resistant to US2-mediated MHC-I loss (green line in histogram) and therefore serves as a functional readout for TRC8 status.
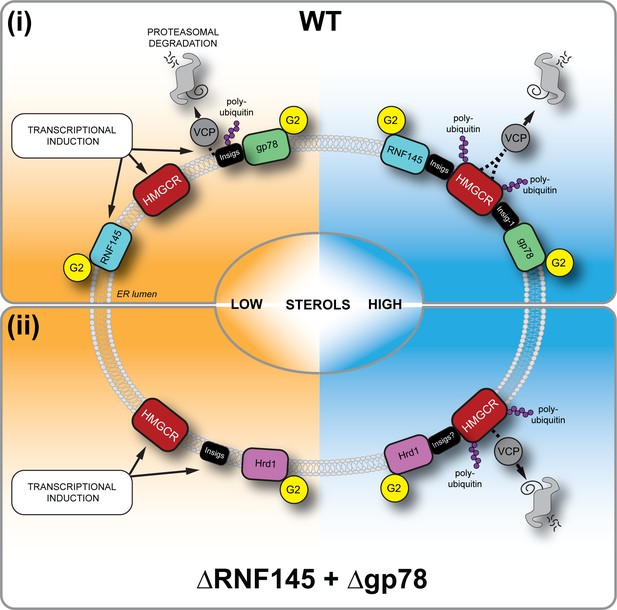
Sterol-induced HMGCR degradation by RNF145, gp78 and Hrd1.
(i) Under sterol-depleted conditions (shaded orange), HMGCR, Insigs and RNF145 are transcriptionally induced leading to accumulation of RNF145 and HMGCR. Insigs are continually turned over by gp78-mediated polyubiquitination, extracted from the membrane by VCP and degraded by the 26S proteasome. HMGCR stability is dramatically increased as it is not engaged by either RNF145, gp78 or their shared E2 ubiquitin conjugating enzyme UBE2G2 (G2). In the presence of sterols (shaded blue), RNF145 and gp78 are recruited to HMGCR in an Insig-assisted fashion, mediating the sterol-accelerated and UBE2G2-dependent degradation of HMGCR by the ubiquitin-proteasome system. Under these conditions both RNF145 and gp78 can independently ubiquitinate HMGCR, which is then extracted from the ER membrane in a VCP-dependent manner. The stoichiometry and make-up of the different Insig complexes within the ER membrane are unknown. (ii) When both RNF145 and gp78 are not available (ΔRNF145 + Δgp78), Hrd1 and UBE2G2 can promote removal of HMGCR in the presence of sterols.
Tables
Candidate genes (- log(p)≥5) identified in a genome-wide CRISPR/Cas9 screen for proteins involved HMGCR degradation.
https://doi.org/10.7554/eLife.40009.007| Gene | Full name | -log(p)* | Function |
|---|---|---|---|
| AUP1 | Ancient Ubiquitous Protein 1 | 4.70 | ERAD |
| EHD1 | EH Domain Containing 1 | 6.50 | Early endosome membrane fusion |
| FAF2 | Fas Associated Factor Family Member 2 | 6.63 | ERAD |
| FER | Tyrosine-protein Kinase Fer | 6.05 | Tyrosine kinase |
| GALNT11 | Polypeptide N-acetylgalactosaminyltransferase 11 | 7.03 | Protein glycosylation |
| INSIG1 | Insulin Induced Gene 1 | 7.50 | Cholesterol metabolism |
| INSIG2 | Insulin Induced Gene 2 | 33.24 | Cholesterol metabolism |
| LDLR | Low Density Lipoprotein Receptor | 11.44 | Cholesterol metabolism |
| PPAP2C | Phospholipid Phosphatase 2 | 5.67 | Glycerolipid synthesis |
| RNF145 | RING Finger Protein 145 | 9.18 | E3 ubiquitin ligase |
| TECR | Trans-2,3-enoyl CoA Reductase | 7.45 | Very-long chain fatty acid synthesis |
| UBE2G2 | Ubiquitin Conjugating Enzyme E2 G2 | 31.14 | E2 ubiquitin conjugating enzyme |
-
*Only statistically significant hits (-log(p)≥5) are shown.
Candidate genes (-log(p)≥5) identified in a ubiquitome CRISPR/Cas9 screen for proteins mediating HMGCR degradation in RNF145-deficient cells.
https://doi.org/10.7554/eLife.40009.013| Gene | Full name | -log(p)* | Function |
|---|---|---|---|
| AMFR | Gp78/Autocrine Motility Factor Receptor | 10.87 | E3 ubiquitin ligase |
| ANKFY1 | Ankyrin Repeat And FYVE Domain Containing 1 | 6.19 | Proposed Rab5 effector |
| FAF2 | Fas Associated Factor Family Member 2 | 5.05 | ERAD |
| NPLOC4 | NPL4 Homolog | 7.63 | Ubiquitin recognition factor |
| RABGEF1 | RAB Guanine Nucleotide Exchange Factor 1 | 8.56 | Nucleotide exchange factor, E3 ubiquitin ligase |
| UBE2G2 | Ubiquitin Conjugating Enzyme E2 G2 | 13.66 | E2 ubiquitin conjugating enzyme |
-
*Only statistically significant hits (-log(p)≥5) are shown.
| Reagent type | Designation | Source | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | anti-gp78 (rabbit polyclonal) | ProteinTech, 16675–1-AP | RRID:AB_2226463 | WB (1:1000) |
| Antibody | anti-ubiquitin (mouse monoclonal) | Life Sensors, VU101 | RRID:AB_2716558 | WB (1:1000) |
| Antibody | anti-V5 tag (mouse monoclonal) | Abcam, ab27671 | RRID:AB_471093 | WB (1:1000) |
| Antibody | anti-Insig-1 (rabbit polyclonal) | Abcam, ab70784 | RRID:AB_1269181 | WB (1:1000) |
| Antibody | anti-Hrd1 (rabbit polyclonal) | Abgent, AP2184a | RRID:AB_2199838 | WB (1:5000) |
| Antibody | anti-TRC8 (rabbit polyclonal) | Santa Cruz, sc-68373 | RRID:AB_2238721 | WB (1:2000) |
| Antibody | anti-HMGCR (mouse monoclonal) | Santa Cruz, sc-271595 | RRID:AB_10650274 | WB (1:1000) |
| Antibody | anti-UBE2G2 (mouse monoclonal) | Santa Cruz Biotechnology, sc-100613 | RRID:AB_1130984 | WB (1:1000) |
| Recombinant DNA reagent | pSpCas9(BB)−2A- Puro V1 (plasmid) | Addgene #48139 | n/a | |
| Recombinant DNA reagent | pSpCas9(BB)−2A-Puro V2 (plasmid) | Addgene #62988 | n/a | |
| Recombinant DNA reagent | pKLV-U6gRNA(BbsI)-PGKpuro2ABFP (plasmid) | Addgene # 50946 | n/a | |
| Recombinant DNA reagent | genome-wide sgRNA library | other | n/a | kind gift from the Bassik lab (Stanford University), PMID: 28474669 |
| Recombinant DNA reagent | ubiquitome sgRNA library | this study | n/a | Generated by Lehner and Nathan labs |
| Peptide, recombinant protein | V5 peptide | Sigma-Aldrich | V7754-4MG | |
| Chemical compound, drug | lipoprotein-deficient serum (LPDS) | Biosera | FB-1001L/100 | |
| Chemical compound, drug | digitonin | Merck | 300410–5 GM | |
| Chemical compound, drug | mevastatin | Sigma-Aldrich | M2537-5MG | |
| Chemical compound, drug | mevalonolactone | Sigma-Aldrich | M4467-1G | |
| Chemical compound, drug | cholesterol | Sigma-Aldrich | C3045-5G | |
| Chemical compound, drug | 25-hydroxycholesterol | Sigma-Aldrich | H1015-10MG | |
| Chemical compound, drug | methyl-β-cyclodextrin (MBCD) | Sigma-Aldrich | 332615–1G | |
| Chemical compound, drug | NMS-873 | Selleckchem | s728501 | |
| Chemical compound, drug | cycloheximide | Sigma-Aldrich | C-7698 | |
| Chemical compound, drug | IgG Sepharose 6 Fast Flow | GE Healthcare | 17-0969-01 | |
| Chemical compound, drug | Protein A-Sepharose | Sigma-Aldrich | P3391-1.5G | |
| Chemical compound, drug | iodoacetamide | Sigma-Aldrich | I1149-5G | |
| Chemical compound, drug | cOmplete protease inhibitor | Roche | 27368400 | |
| Chemical compound, drug | phenylmethyl sulfonyl fluoride (PMSF) | Roche | 20039220 | |
| Chemical compound, drug | N-ethylmaleimide (NEM) | Sigma-Aldrich | E3876-5G | |
| Chemical compound, drug | puromycin | Cayman Chemicals | 13884 | |
| Chemical compound, drug | hygromycin B | Invitrogen | 10687010 |
Additional files
-
Supplementary file 1
sgRNA sequences and genes targeted by the CRISPR/Cas9 ubiquitome library.
- https://doi.org/10.7554/eLife.40009.026
-
Supplementary file 2
sgRNA sequences for generation of knockout cell lines.
- https://doi.org/10.7554/eLife.40009.027
-
Supplementary file 3
Genetically modified cell lines used in this study.
- https://doi.org/10.7554/eLife.40009.028
-
Supplementary file 4
Primers used in CRISPR/Cas9 screens.
- https://doi.org/10.7554/eLife.40009.029
-
Supplementary file 5
Primer sequences used for qPCR.
- https://doi.org/10.7554/eLife.40009.030
-
Transparent reporting form
- https://doi.org/10.7554/eLife.40009.031





