A putative origin of the insect chemosensory receptor superfamily in the last common eukaryotic ancestor
Figures
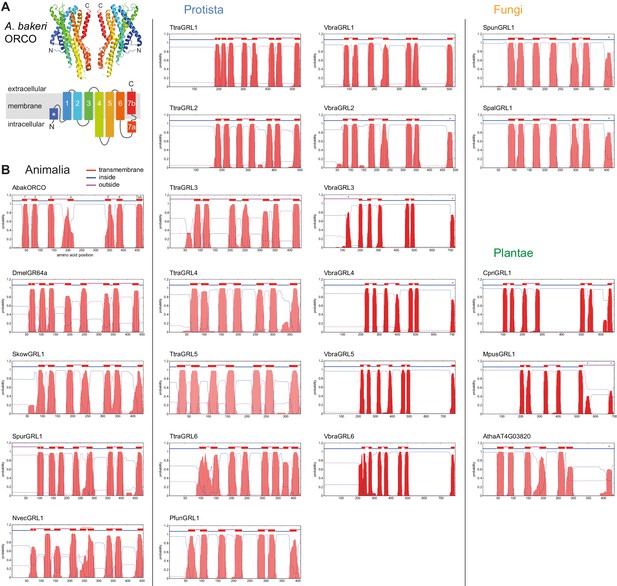
Transmembrane topology predictions of GRLs.
(A) Top: cryo-EM structure of Apocrypta bakeri ORCO (AbakORCO) (PDB 6C70 [Butterwick et al., 2018]); only two subunits of the homotetrameric structure are visualized. Bottom: Schematic of the membrane topology of AbakORCO (adapted from Butterwick et al., 2018), colored as in the cryo-EM structure. The white asterisk marks a helical segment that forms part of a membrane re-entrant loop in the N-terminal region. TM domain seven is divided into a cytoplasmic segment (7a) and a membrane-spanning segment (7b). (B) TM domain and topology predictions of the previously described and newly-recognized GRLs and DUF3537 proteins (Dmel, Drosophila melanogaster; Skow, Saccoglossus kowalevskii; Spur, Strongylocentrotus purpuratus; Nvec, Nematostella vectensis; Atha, Arabidopsis thaliana; see Table 1 for other species abbreviations and sequence accessions). Each plot represents the posterior probabilities of transmembrane helix and inside/outside cellular location along the protein sequence, adapted from the output of TMHMM Server v2 (Krogh et al., 2001). In several sequences an extra transmembrane segment near the N-terminus is predicted (marked by a white asterisk in the N-best prediction above the plot); this may represent the re-entrant loop helical region observed in ORCO, rather than a transmembrane region; in at least one case (SpurGRL1) the designation of this region as a TM domain, leads to an atypical (and presumably incorrect) prediction of an extracellular N-terminus. Conversely, in a subset of proteins individual TM domains are not predicted (notably TM7, black asterisks above the N-best plot), which is likely due to subthreshold predictions for TM domainsin these regions. In NvecGRL1, the long TM4 helix (which projects into the intracellular space in ORCO [Butterwick et al., 2018]) is mis-predicted as two TM domains (dashed red line). Independent membrane topology predictions for unicellular species’ GRLs were obtained using TOPCONs (Supplementary file 2), with largely consistent results.
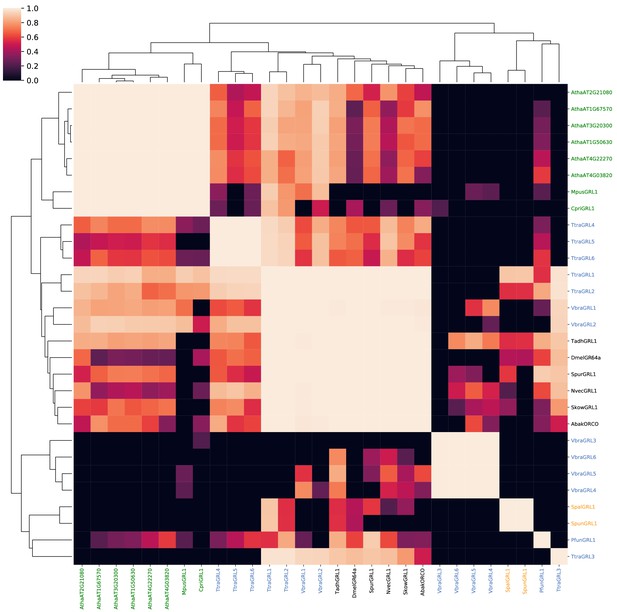
Probabilities of alignments of HMMs of known and candidate GRLs and DUF3537 proteins.
A probability similarity matrix representing the quality of pairwise alignments of the HMMs constructed from the protein sequences indicated in the corresponding rows and columns. The similarity matrix was clustered over its rows and columns using UPGMA hierarchical clustering.
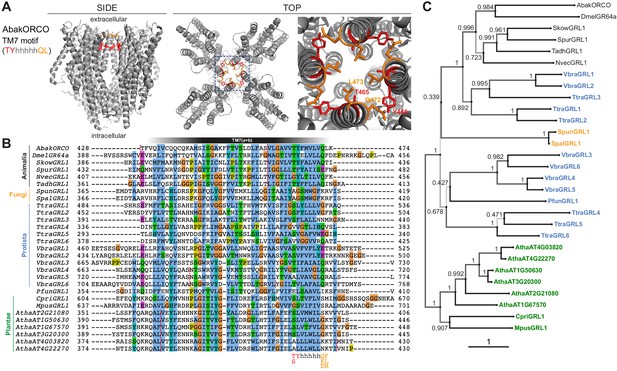
Conservation and divergence in TM7 features and GRL phylogeny.
(A) Side and top views of the cryo-EM structure of the ORCO homotetramer (Butterwick et al., 2018), in which the TM7 motif amino acid side chains are shown in stick format and colored red or orange. The region in the dashed blue box, representing the extracellular entrance to the ion channel pore, is shown in a magnified view on the far right. (B) Multiple sequence alignment of the C-terminal region (encompassing TM7) of unicellular eukaryotic GRLs and selected animal GRLs and plant DUF3537 proteins. Tadh, Trichoplax adhaerens; other species abbreviations are defined in Figure 1 and Table 1. The TM7 motif consensus amino acids (and conservative substitutions) are indicated below the alignment; h indicates a hydrophobic amino acid. Red dashed lines on the alignment indicate positions of predicted introns within the corresponding transcripts. Intron locations are generally conserved within sequences from different Kingdoms, but not between Kingdoms; many Protista sequences do not have introns in this region. (C) Maximum likelihood phylogenetic tree of unicellular eukaryotic GRLs, and selected animal GRLs and plant DUF3537 proteins, with aBayes branch support values. Although the tree is represented as rooted, the rooting is highly uncertain. Protein labels are in black for animals, orange for fungi, blue for protists and green for plants. The scale bar represents one substitution per site.

Alignment of GRL superfamily members.
Multiple sequence alignment of the GRL proteins illustrated in Figure 1. The approximate positions of the TM domains and the N-terminal re-entrant loop (asterisk) are indicated above the alignment.
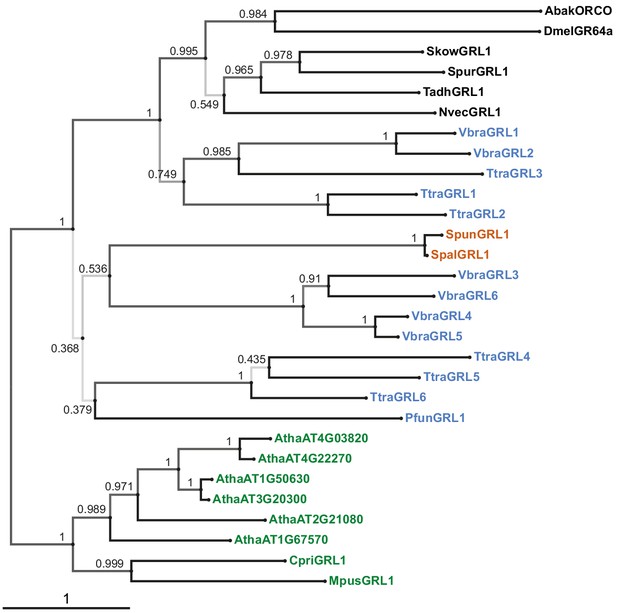
Phylogenetic tree derived from a trimmed multiprotein alignment.
Maximum likelihood phylogenetic tree of unicellular eukaryotic GRLs, and selected animal GRLs and plant DUF3537 proteins, with aBayes branch support values. Although the tree is represented as rooted, the rooting is highly uncertain. Protein labels are in black for animals, orange for fungi, blue for protists and green for plants. The scale bar represents one substitution per site.
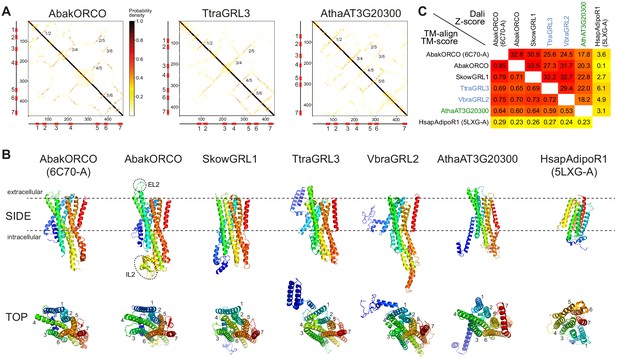
Ab initio structural predictions of GRLs and DUF3537 proteins.
(A) Inter-residue contact maps from trRosetta analysis of the indicated proteins. The axes represent the indices along the primary sequence; the positions of the predicted TM domains are shown in the schematics. The representation is mirror-symmetric along the diagonal; in one half ‘lines’ of contacts perpendicular to the diagonal of the map support the existence of anti-parallel alpha-helical transmembrane packing arrangements. Most pairs of predicted anti-parallel TMs are conserved across the proteins, despite variation in the length of loops between TM domains, supporting a globally similar packing of TM helices. The output of trRosetta analyses for these and other proteins is summarized in Supplementary file 7 and complete datasets are provided in the Dryad repository (doi:10.5061/dryad.s7h44j15f). (B) Side and top views of experimentally-determined (AbakORCO (PDB 6C70 chain A)) and Homo sapiens Adiponectin Receptor 1 (HsapAdipoR1; PDB 5LXG chain A [Vasiliauskaité-Brooks et al., 2017]) or the top trRosetta protein model of GRL and DUF3537 proteins. All GRL/DUF3537 proteins have a similar predicted global packing of TM domains (which is particularly evident in the top view in which the seven TM domains are labelled), despite variation in lengths of the loops and N-terminal regions (colored in dark blue). By contrast, HsapAdipoR1 has a fundamentally different arrangement of TM domains. The dashed ovals on the AbakORCO model highlight the extracellular loop 2 (EL2) and intracellular loop 2 (IL2) regions that were not visualized in the ORCO cryo-EM structure (Butterwick et al., 2018). (C) Quantitative pairwise comparisons of the structures shown in (B) using TM-align (Zhang and Skolnick, 2005) and Dali (Holm and Rosenström, 2010). TM-scores of 0.0–0.30 indicate random structural similarity; TM-scores of 0.5–1.00 indicate that the two proteins adopt generally the same fold (1.00 represents a perfect match). Dali Z-scores of <2 indicate spurious similarity. In both cases, these quantitative cut-offs are not stringent, and must be used as a guide in combination with other criteria (e.g., evidence for homology based upon primary sequence comparisons). The two half-matrices are colored using different scales.
Tables
Candidate GRLs in unicellular eukaryotes.
Protein sequences are provided in Supplementary file 1. Protein nomenclature is provisional and does not imply orthology between species.
| Kingdom | Phylum | Species | Isolate | Alternative name | Common name | Provisional protein name | Accession/version |
|---|---|---|---|---|---|---|---|
| Fungi | Chytridiomycota | Spizellomyces punctatus | DAOM BR117 | chytrid fungus | SpunGRL1 | XP_016607089.1 | |
| Spizellomyces palustris | CBS 455.65 | Phlyctochytrium palustre | chytrid fungus | SpalGRL1 | TPX68946.1 | ||
| Protista | Amoebozoa | Protostelium aurantium var. fungivorum | - | Planoprotostelium fungivorum | - | PfunGRL1 | PRP89608.1 |
| Apusozoa | Thecamonas trahens | ATCC 50062 | Amastigomonas trahens | zooflagellate | TtraGRL1 | XP_013761079.1 | |
| TtraGRL2 | XP_013753662.1 | ||||||
| TtraGRL3 | XP_013759733.1 (trimmed) | ||||||
| TtraGRL4 | XP_013759396.1 | ||||||
| TtraGRL5 | XP_013757274.1 | ||||||
| TtraGRL6 | XP_013755387.1 | ||||||
| Incertae sedis/Chromerdia (superphylum: Alveolata) | Vitrella brassicaformis | CCMP3315 | - | chromerid | VbraGRL1 | CEM13019.1 | |
| VbraGRL2 | CEL93132.1 | ||||||
| VbraGRL3 | CEM19221.1 | ||||||
| VbraGRL4 | CEM01650.1 | ||||||
| VbraGRL5 | CEM10760.1 | ||||||
| VbraGRL6 | CEM25255.1 | ||||||
| Plantae | Chlorophyta | Chloropicon primus | - | CpriGRL1 | QDZ19318.1 | ||
| Micromonas pusilla | CCMP1545 | Chromulina pusilla | MpusGRL1 | XP_003054778.1 |
Additional files
-
Supplementary file 1
Protein sequences of candidate unicellular eukaryotic GRLs.
Provisional protein nomenclature (as used in the figures) is indicated in the header of each sequence. Note that these names do not imply orthology between species. Manual corrections to sequences are also noted in the header (e.g., in TtraGRL3 a large C-terminal region was removed as this is likely due to an incorrect merging of exons of adjacent genes that are separated by a gap in the genomic sequence assembly).
- https://cdn.elifesciences.org/articles/62507/elife-62507-supp1-v2.txt.zip
-
Supplementary file 2
TOPCONs analysis output of candidate unicellular eukaryotic GRLs.
- https://cdn.elifesciences.org/articles/62507/elife-62507-supp2-v2.zip
-
Supplementary file 3
Sequences retrieved through the HMM searches.
Each row contains a separate hit, with identifier, probability, length, query, and score.
- https://cdn.elifesciences.org/articles/62507/elife-62507-supp3-v2.csv.zip
-
Supplementary file 4
Code to generate Figure 1—figure supplement 1 (matrix of pairwise HMM alignment probabilities).
The Python code is provided as a Jupyter notebook in HTML format, and includes the specific arguments used to run the HHsuite tools.
- https://cdn.elifesciences.org/articles/62507/elife-62507-supp4-v2.zip
-
Supplementary file 5
Multiprotein alignment of GRLs and DUF3537 proteins.
- https://cdn.elifesciences.org/articles/62507/elife-62507-supp5-v2.txt.zip
-
Supplementary file 6
Trimmed multiprotein alignment of GRLs and DUF3537 proteins.
- https://cdn.elifesciences.org/articles/62507/elife-62507-supp6-v2.txt.zip
-
Supplementary file 7
Ab initio protein modeling of GRLs and DUF3537 proteins.
Results from trRosetta and RaptorX analyses with the indicated protein queries. Multiple sequence alignments (MSAs) were built automatically with the indicated numbers of sequences. In several cases, insufficient sequences were aligned, leading to inadequate data for prediction of inter-residue contacts. For these proteins, the ‘estimated TM-scores’ of the trRosetta models are commensurately low (scores < 0.17 are likely to reflect spurious protein structural models [Yang et al., 2020; Zhang and Skolnick, 2004]) and further analysis was not pursued (as indicated by the grey cells). For the other proteins, the top hit and corresponding Z-score from Dali searches of the full Protein Data Bank (PDB) with the top-predicted model are shown. The top model for RaptorX output was defined as that with the lowest ‘estimated RMSD’ (root mean-squared deviation, i.e., the estimated average distance deviation (in Å) of the model from the real structure). For almost all models, individual chains of the AbakORCO homotetramer (A-D) were retrieved as top hits, with a much higher Z-score than the next, non-ORCO hit. TtraGRL4 and TtraGRL5 models were built using MSAs containing only a subset of plant DUF3537 proteins. Although the trRosetta estimated TM-scores were above the threshold (i.e.,>0.17), the retrieved Dali top hits did not have stand-out Z-scores and are likely to be spurious (DIABLO is a HECT-type E3 ligase and PLECTIN is a cytoskeletal protein); here the Z-score of the AbakORCO hit is also given. Dali searches with several of the RaptorX models of the plant proteins identified de novo designed (i.e., artificial) proteins or completely unrelated molecules as the top hit, but AbakORCO was usually also retrieved, with a lower Z-score, as indicated. Full output of trRosetta and RaptorX analyses and Dali searches are provided in the Dryad repository (doi:10.5061/dryad.s7h44j15f).
- https://cdn.elifesciences.org/articles/62507/elife-62507-supp7-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/62507/elife-62507-transrepform-v2.docx





