Volatile DMNT directly protects plants against Plutella xylostella by disrupting the peritrophic matrix barrier in insect midgut
Figures
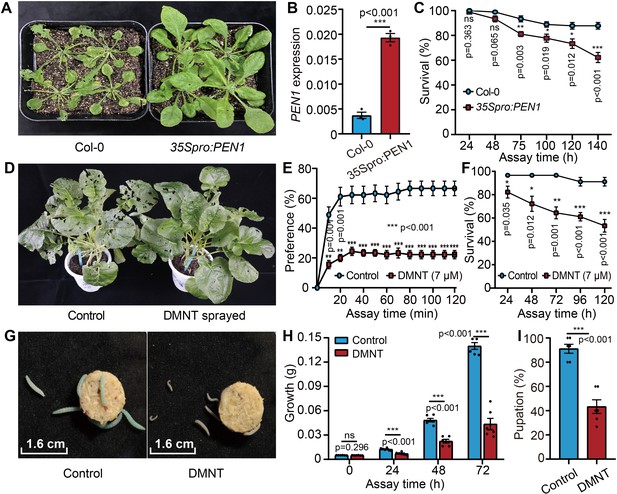
(3E)-4,8-Dimethyl-1,3,7-nonatriene (DMNT) repels and kills P. xylostella larvae.
(A) Pentacyclic triterpene synthase 1 (PEN1) overexpression in A. thaliana results in high resistance to P. xylostella infestation. PEN1 encodes the key enzyme responsible for DMNT biosynthesis in Arabidopsis plants. (B) PEN1 is overexpressed in 35Spro:PEN1 transgenic Arabidopsis plants. (C) 35Spro:PEN1 transgenic Arabidopsis plants cause lower survival of P. xylostella than wild type control. (D) B. napus plants sprayed with DMNT show strong resistance to P. xylostella larvae. (E) P. xylostella larvae can sense and are repelled by DMNT. The preference test system is illustrated in Figure 1—figure supplement 2. (F) DMNT treatment significantly lowers the survival of P. xylostella larvae. (G) Phenotypic comparison between DMNT-fed larvae and control larvae (treated with the DMNT solvent paraffin oil). (H) Growth inhibition of P. xylostella larvae by DMNT. (I) Reduced pupation of P. xylostella larvae by DMNT treatment. Error bars represent the standard error of six independent biological replicates, with 15 larvae per replicate. Asterisks indicate significant differences (*p<0.05, **p<0.01, ***p<0.001, ns, not significant; two-tailed unpaired t-test).
-
Figure 1—source data 1
(3E)-4,8-Dimethyl-1,3,7-nonatriene (DMNT) repels and kills P. xylostella larvae.
- https://cdn.elifesciences.org/articles/63938/elife-63938-fig1-data1-v2.xlsx
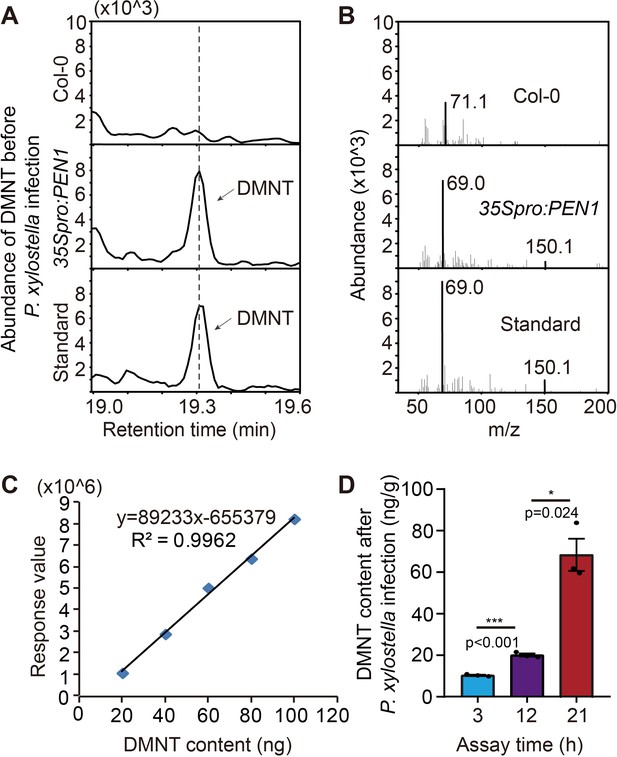
(3E)-4,8-Dimethyl-1,3,7-nonatriene (DMNT) is enriched in 35Spro:PEN1 transgenic A. thaliana plants and can be highly induced by P. xylostella infestation.
(A) Gas chromatography-mass spectrometry analysis of DMNT in Col-0 and 35Spro:PEN1 transgenic A. thaliana plants. The DMNT standard is shown at the bottom. (B) Ionogram traces of DMNT captured from Col-0 and 35Spro:PEN1 transgenic plants. (C) A linear standard curve of DMNT quantification, which was dissolved in methanol. (D) DMNT in 35Spro:PEN1 transgenic A. thaliana plants can be continuously induced by P. xylostella infestation for 3–21 hr. Asterisks in (D) indicate significant differences (*p<0.05, ***p<0.001; two-tailed unpaired t-test).
-
Figure 1—figure supplement 1—source data 1
(3E)-4,8-Dimethyl-1,3,7-nonatriene (DMNT) is enriched in 35Spro:PEN1 transgenic A. thaliana plants and can be highly induced by P. xylostella infestation.
- https://cdn.elifesciences.org/articles/63938/elife-63938-fig1-figsupp1-data1-v2.xlsx
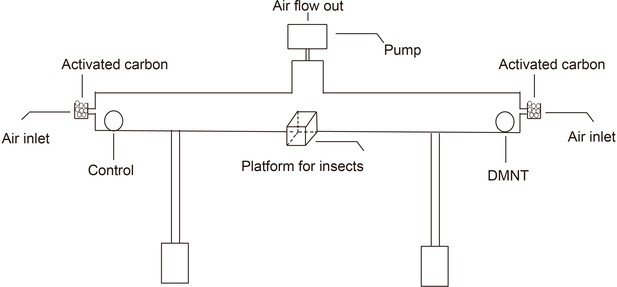
A schematic representation of the principle behind the insect choice test system.
The horizontal box indicates the glass test tube. Equal amounts of activated carbon are placed at both ends of the test tube to avoid air contamination. The air flow is generated by an air pump. In all experiments, (3E)-4,8-dimethyl-1,3,7-nonatriene (DMNT) and control samples are placed on either end of the glass test tube, while 15 P. xylostella larvae are put on the platform located in the center of the test tube for the choice test.
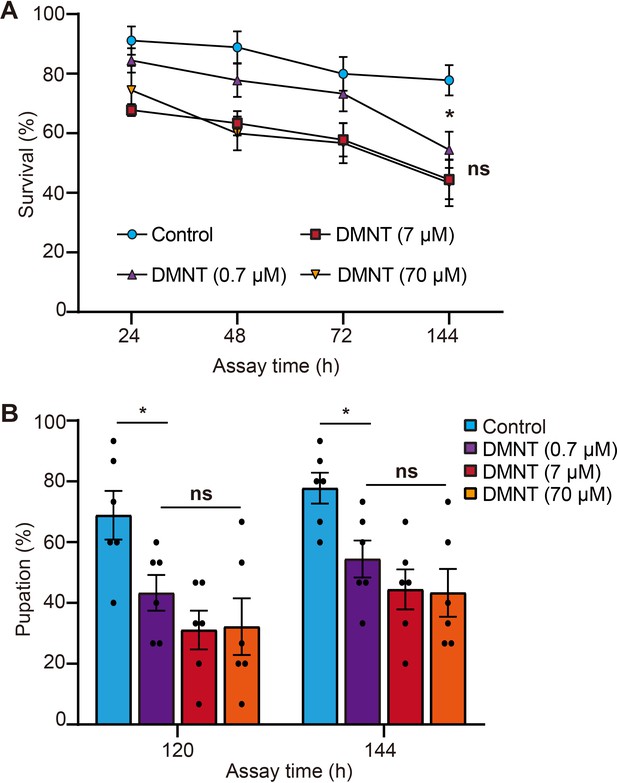
(3E)-4,8-Dimethyl-1,3,7-nonatriene (DMNT) shows a slight dosage-dependent influence on the survival and pupation rates of P. xylostella larvae.
(A) DMNT shows no dosage-dependent effects on the end of survival or pupation (B) of P. xylostella larvae. Error bars in (A, B) represent the standard error of six biological replicates, with 15 larvae per replicate. Asterisks indicate significant differences (*p<0.05; ns, not significant; two-tailed unpaired t-test).
-
Figure 1—figure supplement 3—source data 1
(3E)-4,8-Dimethyl-1,3,7-nonatriene (DMNT) shows a slight dosage-dependent influence on the survival and pupation rates of P. xylostella larvae.
- https://cdn.elifesciences.org/articles/63938/elife-63938-fig1-figsupp3-data1-v2.xlsx
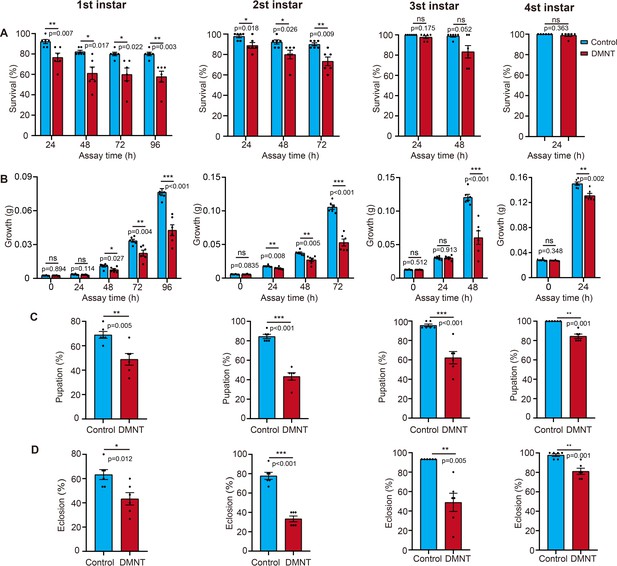
(3E)-4,8-Dimethyl-1,3,7-nonatriene (DMNT) treatment affects P. xylostella larvae of different ages.
First- to fourth-instar P. xylostella larvae were tested. (A) Survival rate. (B) Body weight. (C) Pupation rate. (D) Eclosion rate. Error bars represent the standard error of six biological replicates, with 15 P. xylostella larvae per replicate. Asterisks indicate significant differences (*p<0.05, **p<0.01, ***p<0.001; ns, not significant; two-tailed unpaired t-test).
-
Figure 1—figure supplement 4—source data 1
(3E)-4,8-Dimethyl-1,3,7-nonatriene (DMNT) treatment affects P. xylostella larvae of different ages.
- https://cdn.elifesciences.org/articles/63938/elife-63938-fig1-figsupp4-data1-v2.xlsx
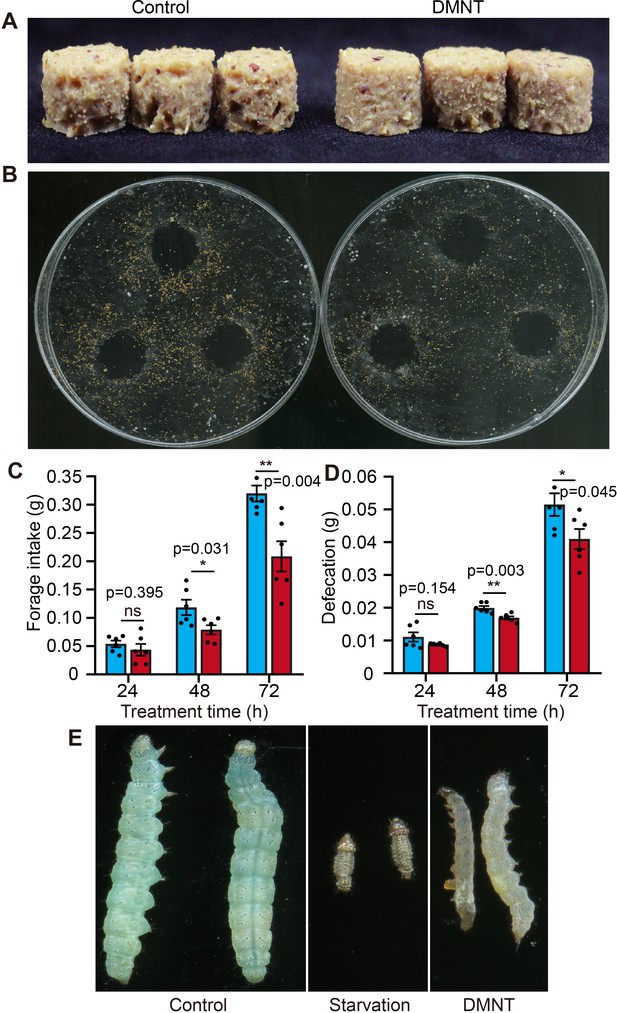
(3E)-4,8-Dimethyl-1,3,7-nonatriene (DMNT) treatment lowers forage intake and defecation of P. xylostella larvae.
(A, C) Reduced forage intake in P. xylostella larvae fed with DMNT. (B, D) Reduction in defecation of DMNT-fed P. xylostella larvae compared to the control. Error bars in (C, D) represent the standard error of six biological replicates, each consisting of 15 P. xylostella larvae. The same amount of forage was used in all assays. (E) The length of DMNT-treated P. xylostella is different from that of starved larvae. Asterisks indicate significant differences (*p<0.05, **p<0.01; ns, not significant; two-tailed unpaired t-test).
-
Figure 1—figure supplement 5—source data 1
(3E)-4,8-Dimethyl-1,3,7-nonatriene (DMNT) treatment lowers forage intake and defecation of P. xylostella larvae.
- https://cdn.elifesciences.org/articles/63938/elife-63938-fig1-figsupp5-data1-v2.xlsx
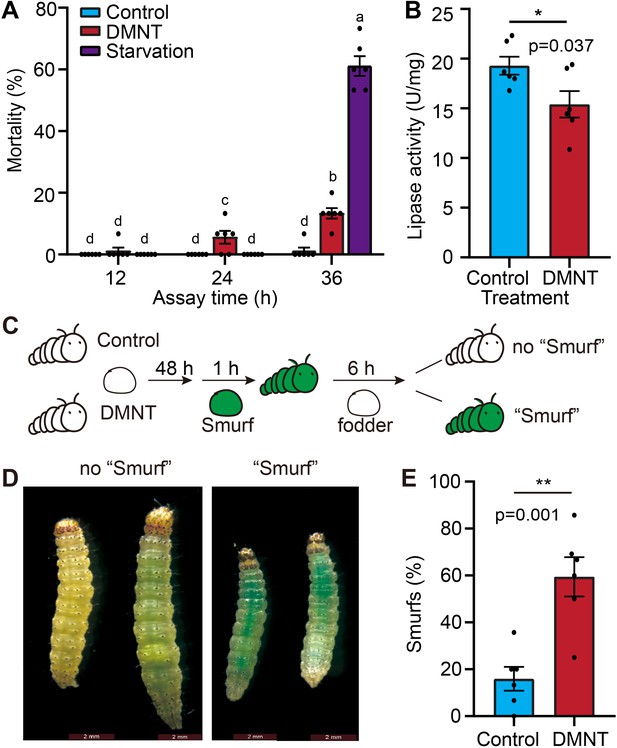
The midgut barrier of P. xylostella larvae is damaged by (3E)-4,8-dimethyl-1,3,7-nonatriene (DMNT) treatment.
(A) Larvae die earlier from treatment with DMNT than from starvation. (B) Decreased lipase activity in the midgut of P. xylostella larvae upon DMNT treatment. (C) The principle of the Smurf test. Larvae are fed with forage loaded with the dye erioglaucine disodium salt for 1 hr before returning to normal forage without dye. After 6 hr, the extent of dye retention is monitored. (D) Representative images of DMNT-treated larvae showing dye retention, as evidenced by their blue appearance, like the Smurf cartoon character. (E) Quantification of results shown in (D). The different letters in (A) indicate a significant difference (one-way ANOVA). Error bars in (B, E) represent the standard error of six independent biological replicates, with 15 larvae per replicate. Asterisks in (B, E) indicate significant differences (*p<0.05, **p<0.01, ns, not significant; two-tailed unpaired t-test).
-
Figure 2—source data 1
The midgut barrier of P. xylostella larvae is damaged by (3E)-4,8-dimethyl-1,3,7-nonatriene (DMNT) treatment.
- https://cdn.elifesciences.org/articles/63938/elife-63938-fig2-data1-v2.xlsx
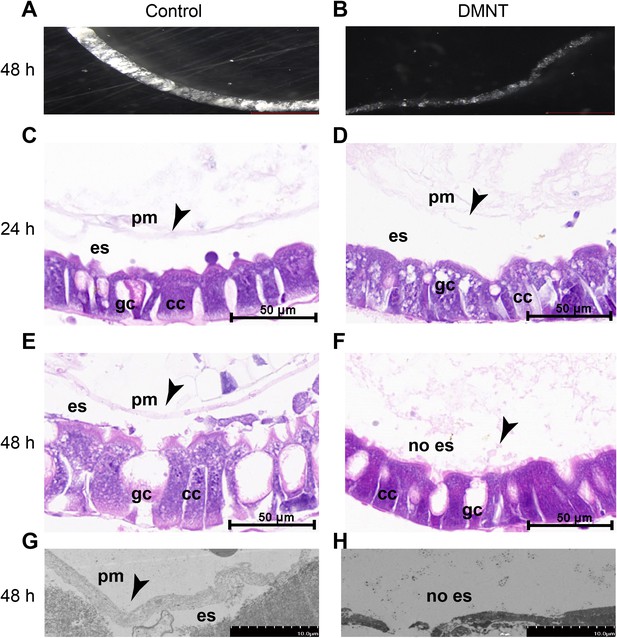
The peritrophic matrix (PM) structure is damaged by (3E)-4,8-dimethyl-1,3,7-nonatriene (DMNT) treatment.
(A, B) PM ultrastructure of control (A) and DMNT-fed (B) P. xylostella larvae for 48 hr. Note the thin and delicate PM in DMNT-fed larvae. (C–F) Transverse section and hematoxylin-eosin staining show damage of the PM by DMNT after exposure for 24 hr (C, D) and 48 hr (E, F). (G, H) The PM is damaged by DMNT feeding, as shown by transmission electron microscopy. Arrowheads indicate the PM. es: delimits the ectoperitrophic space; gc: goblet cells; cc: columnar cells.
-
Figure 3—source data 1
The peritrophic matrix (PM) structure is damaged by (3E)-4,8-dimethyl-1,3,7-nonatriene (DMNT) treatment.
- https://cdn.elifesciences.org/articles/63938/elife-63938-fig3-data1-v2.xlsx
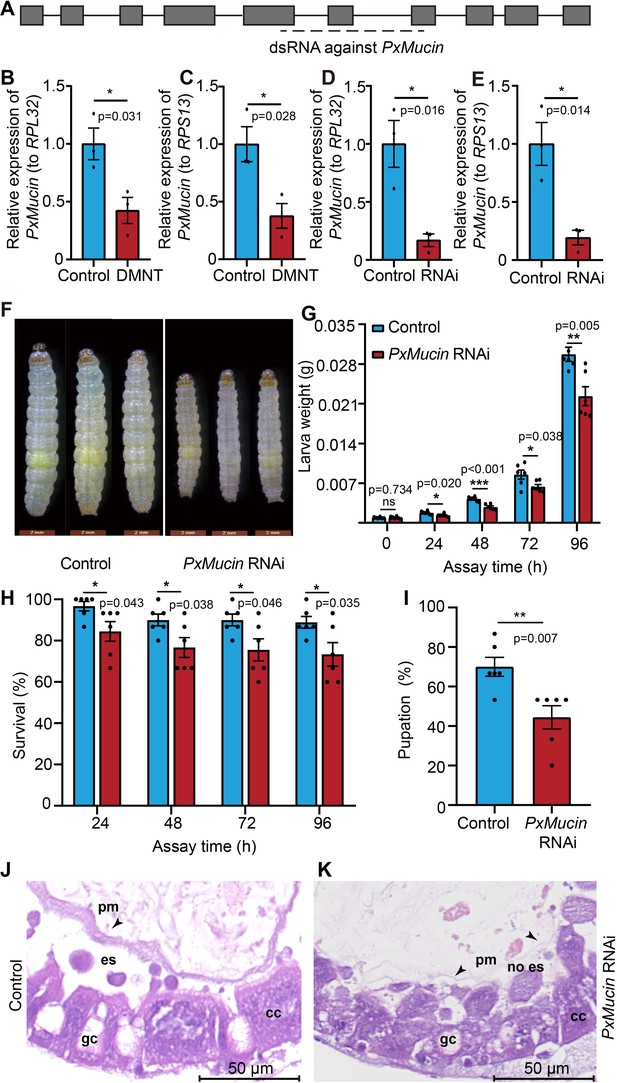
(3E)-4,8-Dimethyl-1,3,7-nonatriene (DMNT) disrupts the peritrophic matrix (PM) structure of P. xylostella larvae via repression of the mucin-like gene PxMucin.
(A) Gene structure of PxMucin. Boxes represent exons, and horizontal lines represent introns. The horizontal dashed line indicates the targeted regions by double-stranded RNA (dsRNA). (B, C) DMNT treatment downregulates the expression of PxMucin. (D, E) Successful RNA interference (RNAi) of PxMucin by dsRNA feeding. (F–I) Larvae fed with dsRNA against PxMucin show impaired body development (F), weight (G), survival (H), and pupation (I) rates. (J, K) PM structure from control larvae (J) and larvae-fed PxMucin dsRNA (K). Note how PxMucin dsRNA feeding phenocopies DMNT treatment. Error bars in (B–E) represent the standard error of three independent biological replicates, with 30 P. xylostella larvae per replicate. Error bars in (G–I) represent the standard error of six independent biological replicates, with 15 P. xylostella larvae per replicate. Asterisks indicate significant differences (*p<0.05, **p<0.01, ***p<0.001, ns, not significant; two-tailed unpaired t-test).
-
Figure 4—source data 1
(3E)-4,8-Dimethyl-1,3,7-nonatriene (DMNT) disrupts the peritrophic matrix (PM) structure of P. xylostella larvae via repression of the mucin-like gene PxMucin.
- https://cdn.elifesciences.org/articles/63938/elife-63938-fig4-data1-v2.xlsx
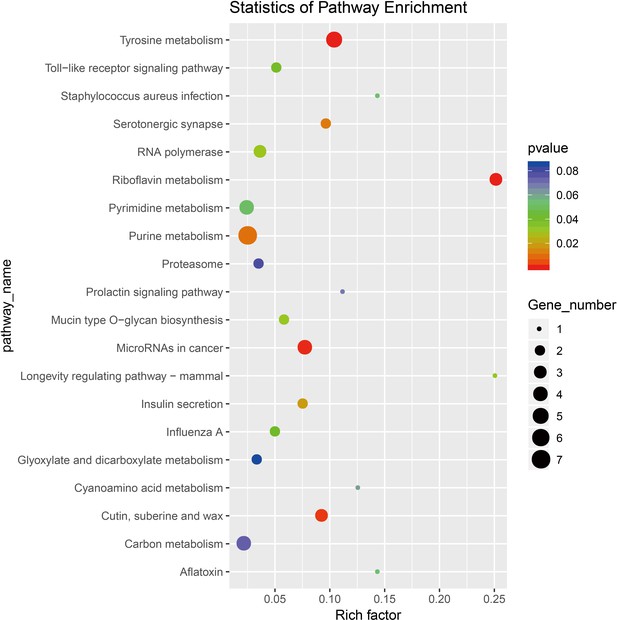
Transcriptomic analysis of the differentially expressed genes in the P. xylostella larva after (3E)-4,8-dimethyl-1,3,7-nonatriene (DMNT) treatment at 12 hr.
Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of the pathway enrichment from the differently expressed genes after DMNT treatment is shown.
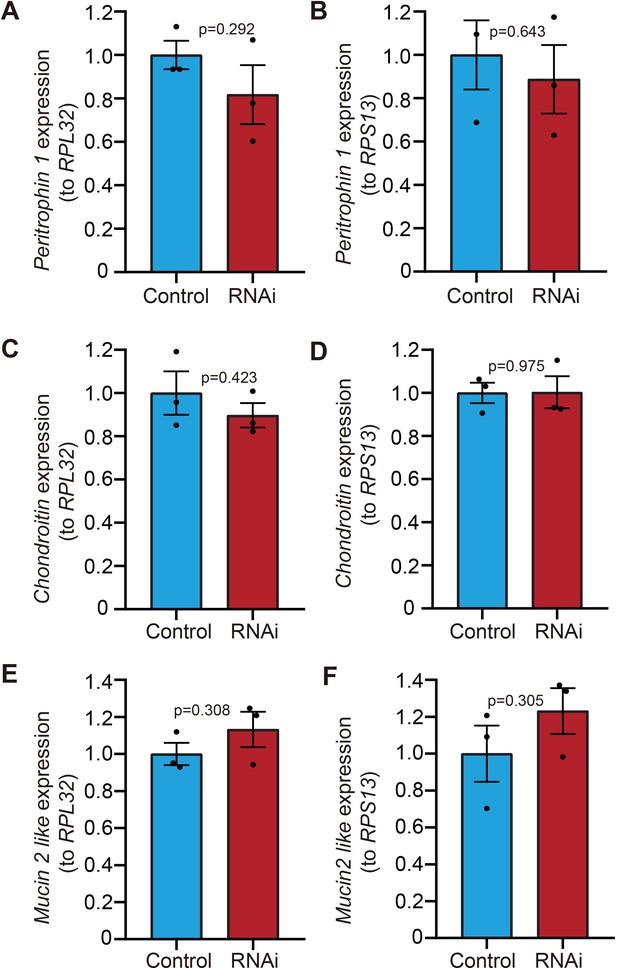
The expression of three PxMucin homologs is not significantly influenced by the double-stranded RNA (dsRNA) treatment.
(A, B) Peritrophin 1 like. (C, D) Chondroitin. (E, F) Mucin 2 like. RPL32 and RPS13 were used as internal control Fu et al., 2013 for qRT-PCR. Error bars represent the standard error of three biological replicates. p values of statistical tests are shown in figures (two-tailed unpaired t-test).
-
Figure 4—figure supplement 2—source data 1
The expression of three PxMucin homologs is not significantly influenced by the double-stranded RNA (dsRNA) treatment.
- https://cdn.elifesciences.org/articles/63938/elife-63938-fig4-figsupp2-data1-v2.xlsx
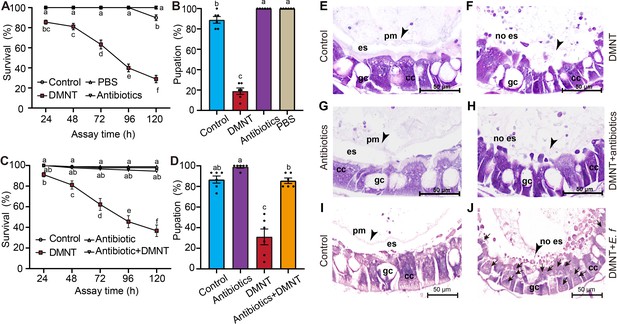
Microbes in the midgut are indispensable for (3E)-4,8-dimethyl-1,3,7-nonatriene (DMNT)-mediated killing of P. xylostella larvae.
(A, B) Removal of the microbiota by antibiotics alone has no influence on larval survival (A) or pupation rates (B). DMNT treatment was used as a control. (C, D) Removal of the microbiota by treatment with antibiotics eliminates the adverse DMNT-mediated effects on the survival (C) and pupation (D) rates of larvae. (E–H) Comparison of peritrophic matrix (PM) structure in control larvae (E), larvae treated with DMNT alone, (F) or in combination with antibiotics (H). Note the disruption of the PM, whereas treatment with the antibiotic cocktail alone has no effects (G). (I, J) Enterococcus faecalis (E. f) can invade midgut cells when the PM is disrupted by DMNT treatment (J), but is restricted within the PM in controls (I). Arrows indicate E. f cells stained with dye. Arrowheads indicate the PM. es: delimits the ectoperitrophic space; gc: goblet cells; cc: columnar cells. The different letters in (A–D) indicate a significant difference (one-way ANOVA).
-
Figure 5—source data 1
Microbes in the midgut are indispensable for (3E)-4,8-dimethyl-1,3,7-nonatriene (DMNT)-mediated killing of P. xylostella larvae.
- https://cdn.elifesciences.org/articles/63938/elife-63938-fig5-data1-v2.xlsx
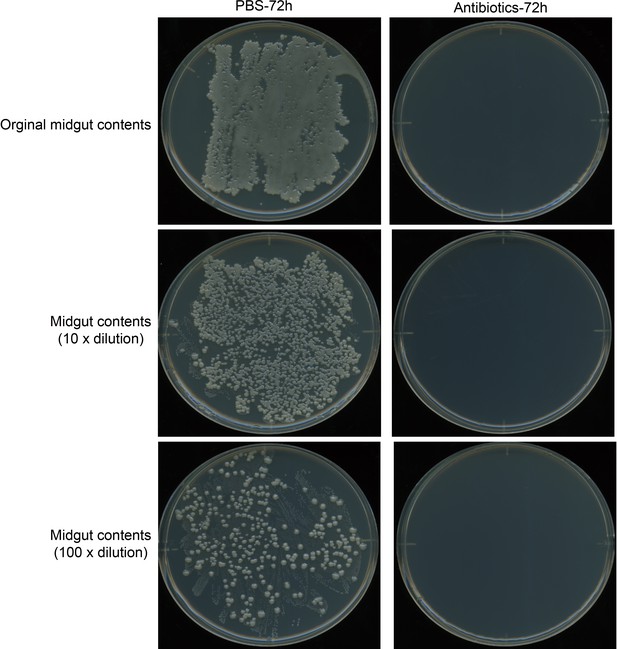
The midgut microbiota is completely killed by the antibiotic cocktail.
Multiple replicates were carried out with the same concentrations of antibiotics added to forage to feed P. xylostella larvae, with similar results. The antibiotic cocktail contains 100 kU/mL penicillin, 100 mg/mL streptomycin, and 50 mg/mL gentamycin.
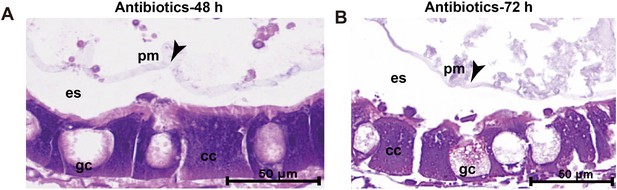
Treatment with antibiotics does not influence the integrity of peritrophic matrix (PM) structures.
(A) 48-hr treatment with antibiotics. (B) 72-hr treatment with antibiotics. Arrowheads indicate the PM. es: delimits the ectoperitrophic space; gc: goblet cells; cc: columnar cells. The antibiotic cocktail contains 100 kU/mL penicillin, 100 mg/mL streptomycin, and 50 mg/mL gentamycin.
-
Figure 5—figure supplement 2—source data 1
Treatment with antibiotics does not influence the integrity of peritrophic matrix (PM) structures.
- https://cdn.elifesciences.org/articles/63938/elife-63938-fig5-figsupp2-data1-v2.xlsx
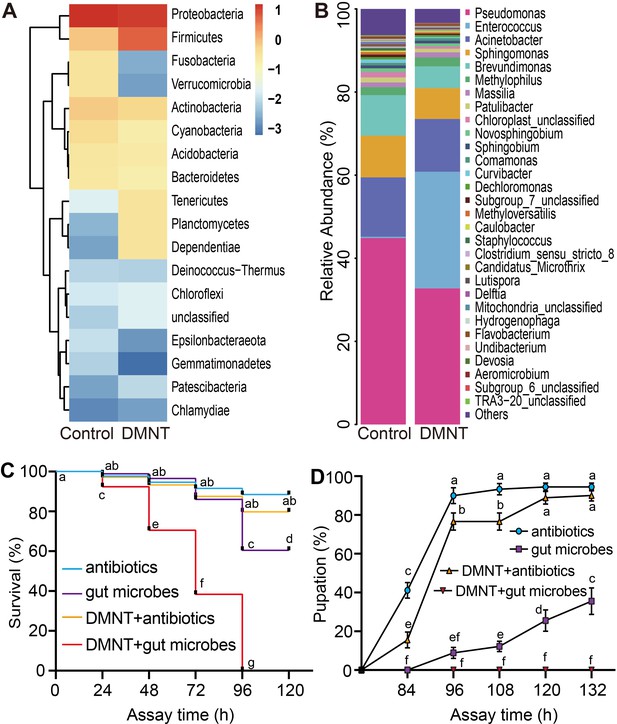
The microbiota composition of P. xylostella larvae midgut is affected by (3E)-4,8-dimethyl-1,3,7-nonatriene (DMNT) treatment.
(A) Relative abundance of 18 phyla in response to DMNT treatment. (B) Effect of DMNT treatment on Enterococcus microbial populations. (C, D) Feeding with gut microbes decreases the survival (C) and pupation (D) rates of larvae treated with DMNT. Error bars represent the standard error of six independent biological replicates, with 15 P. xylostella larvae each. The different letters in (C, D) indicate a significant difference (one-way ANOVA).
-
Figure 6—source data 1
The microbiota composition of P. xylostella larvae midgut is affected by (3E)-4,8-dimethyl-1,3,7-nonatriene (DMNT) treatment.
- https://cdn.elifesciences.org/articles/63938/elife-63938-fig6-data1-v2.xlsx
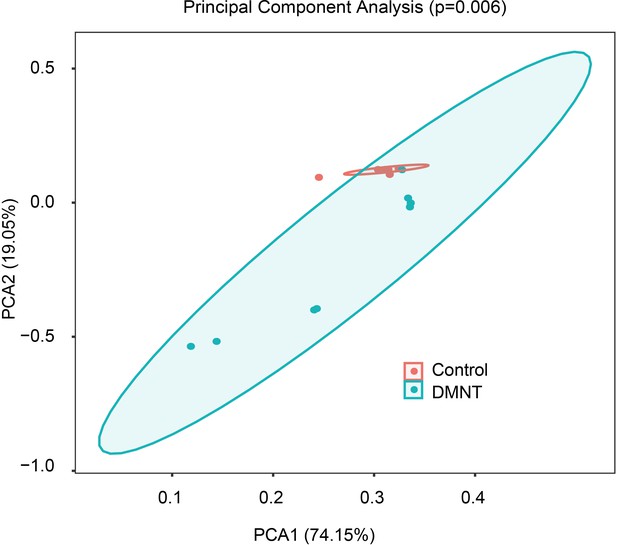
(3E)-4,8-Dimethyl-1,3,7-nonatriene (DMNT) affects the abundance of microbiota populations in the midgut of P. xylostella larvae.
Principal component analysis of the 16S rDNA sequencing results. DMNT-treated samples cluster separately from control samples (p=0.006).
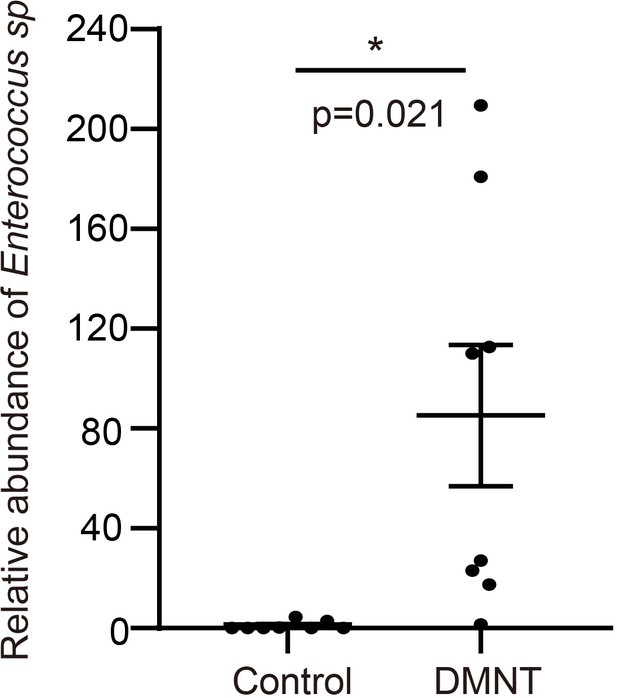
Enterococcus sp. was enriched around 85-fold in (3E)-4,8-dimethyl-1,3,7-nonatriene (DMNT)-treated larvae than the control larvae.
Asterisks indicate significant differences (*p<0.05; two-tailed unpaired t-test).
-
Figure 6—figure supplement 2—source data 1
Enterococcus sp. was enriched around 85-fold in (3E)-4,8-dimethyl-1,3,7-nonatriene (DMNT)-treated larvae than the control larvae.
- https://cdn.elifesciences.org/articles/63938/elife-63938-fig6-figsupp2-data1-v2.xlsx
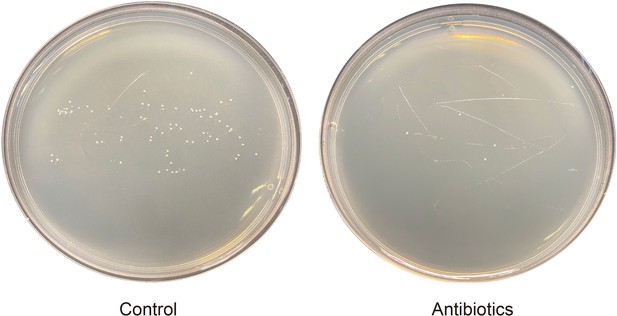
The gut microbiota from second-instar P. xylostella larvae were nearly killed by the antibiotic cocktail.
The antibiotic cocktail contains 100 kU/mL penicillin, 100 mg/mL streptomycin, and 50 mg/mL gentamycin.
-
Figure 6—figure supplement 3—source data 1
The gut microbiota from second-instar P. xylostella larvae were nearly killed by the antibiotic cocktail.
- https://cdn.elifesciences.org/articles/63938/elife-63938-fig6-figsupp3-data1-v2.xlsx
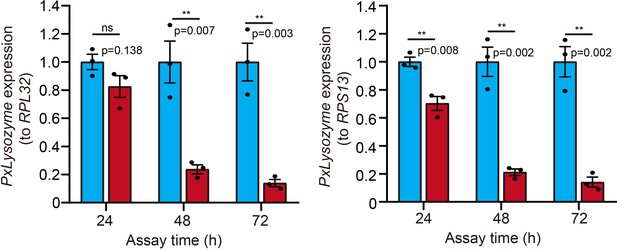
PxLysozyme expression was downregulated from 48 to 72 hr.
Error bars represent the standard error of three independent biological replicates, with 30 P. xylostella larvae per replicate. Asterisks indicate significant differences (**p<0.01, ns, not significant; two-tailed unpaired t-test).
-
Figure 6—figure supplement 4—source data 1
PxLysozyme expression was downregulated from 48 to 72 hr.
- https://cdn.elifesciences.org/articles/63938/elife-63938-fig6-figsupp4-data1-v2.xlsx
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Arabidopsis thaliana) | PEN1 | Sohrabi et al., 2015 | At4g15340 (Tair) | 35Spro:PEN1-Nos construction |
| Gene (Plutella xylostella) | PxMucin | Sarauer et al., 2003; Israni and Rajam, 2017 | LOC105381635(NCBI) | qRT-PCR assay |
| Strain, strain background (Enterococcus sp.) | Enterococcus faecalis (E. f) | This paper | ATCC29212 (http://www.bncc.org.cn/) | Gram staining |
| Commercial assay or kit | T7 RiboMAX express RNAi synthesis kit | This paper | Promega, catalog number P1700 | Double-stranded RNA synthesis |
| Commercial assay or kit | Lipase activity | This paper | Jiancheng Company (http://www.njjcbio.com, catalog: A054-1-1) | Lipase enzyme activity assay |
| Chemical compound, drug | Dye erioglaucine disodium salt | Amcheslavsky et al., 2009; Rera et al., 2012; He et al., 2017 | Sigma-Aldrich, cas: 3844-45-9, molecular weight: 792.85, color index number 42090 | ‘Smurf’ treatment |
| Chemical compound, drug | Antibiotic cocktail | Rodgers et al., 2017 | Solarbio, P1410 | Killing gut microbes |
| Chemical compound, drug | DMNT | Huang and Yang, 2007 | Chemical synthesis of DMNT | |
| Software, algorithm | SPSS | SPSS | RRID:SCR_002865 | statistical analyses |
| Other | Polydimethylsiloxane solid phase micro-extraction (SPME) fibers | Sohrabi et al., 2015 | 100 μm, DVB/CAR/PDMS, Supelco, Inc, Bellefonte, PA | Collection of volatile compounds |
Additional files
-
Supplementary file 1
Differentially expressed genes from RNA-seq.
- https://cdn.elifesciences.org/articles/63938/elife-63938-supp1-v2.xlsx
-
Supplementary file 2
Primers used in this study.
- https://cdn.elifesciences.org/articles/63938/elife-63938-supp2-v2.doc
-
Supplementary file 3
Abundance of major Operational Taxonomic Units (OTUs).
- https://cdn.elifesciences.org/articles/63938/elife-63938-supp3-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/63938/elife-63938-transrepform-v2.docx





