Recognition of discrete export signals in early flagellar subunits during bacterial type III secretion
Abstract
Type III Secretion Systems (T3SS) deliver subunits from the bacterial cytosol to nascent cell surface flagella. Early flagellar subunits that form the rod and hook substructures are unchaperoned and contain their own export signals. A gate recognition motif (GRM) docks them at the FlhBc component of the FlhAB-FliPQR export gate, but the gate must then be opened and subunits must be unfolded to pass through the flagellar channel. This induced us to seek further signals on the subunits. Here, we identify a second signal at the extreme N-terminus of flagellar rod and hook subunits and determine that key to the signal is its hydrophobicity. We show that the two export signal elements are recognised separately and sequentially, as the N-terminal signal is recognised by the flagellar export machinery only after subunits have docked at FlhBC via the GRM. The position of the N-terminal hydrophobic signal in the subunit sequence relative to the GRM appeared to be important, as a FlgD deletion variant (FlgDshort), in which the distance between the N-terminal signal and the GRM was shortened, ‘stalled’ at the export machinery and was not exported. The attenuation of motility caused by FlgDshort was suppressed by mutations that destabilised the closed conformation of the FlhAB-FliPQR export gate, suggesting that the hydrophobic N-terminal signal might trigger opening of the flagellar export gate.
Editor's evaluation
Herein, the authors show that the flagellar Type III Secretion System recognize sequentially two discrete export signals, 1. initial docking and 2. subsequent opening of the export gate, for bacterial flagella biogenesis. This important and elegantly designed study elucidates a key step in solving the long-standing question of how export substrates are recognized by the type III secretion system.
https://doi.org/10.7554/eLife.66264.sa0Introduction
Type III Secretion Systems (T3SS) are multi-component molecular machines that deliver protein cargo from the bacterial cytosol either to their site of assembly in cell surface flagella or virulence factor injectisomes, or directly to their site of action in eukaryotic target cells or the extracellular environment (Evans et al., 2014a; Deng et al., 2017; Büttner and He, 2009; Konkel et al., 2004; Dongre et al., 2018). The flagellar T3SS (fT3SS) directs the export of thousands of structural subunits required for the assembly and operation of flagella, rotary nanomotors for cell motility that extend from the bacterial cell surface (Evans et al., 2014a; Evans et al., 2014b). Newly synthesised subunits of the flagellar rod, hook and filament are targeted to the fT3SS, where they are unfolded and translocated across the cell membrane, powered by the protonmotive force and ATP hydrolysis, into an external export channel that spans the length of the nascent flagellum (Minamino and Namba, 2008; Paul et al., 2008). During flagellum biogenesis, when the rod/hook structure reaches its mature length, the fT3SS switches export specificity from recognition of ‘early’ rod/hook subunits to ‘late’ subunits for filament assembly (Williams et al., 1996; Fraser et al., 2003). This means that early and late flagellar subunits must be differentiated by the fT3SS machinery to ensure that they are exported at the correct stage of flagellum biogenesis. This is achieved, in part, by targeting subunits to the export machinery at the right time using a combination of export signals in the subunit mRNA and/or polypeptide. T3SS substrates contain N-terminal signals for targeting to the export machinery, however they do not share a common peptide sequence (Kuwajima et al., 1989; Minamino and Macnab, 1999; Kornacker and Newton, 1994; Evans et al., 2013; Végh et al., 2006). In addition, some substrates are piloted to the T3SS machinery by specific chaperones (Wattiau et al., 1994; Fraser et al., 1999; Thomas et al., 2004; Akeda and Galán, 2005; Bange et al., 2010).
The core export components of the fT3SS are evolutionarily related to those of the virulence injectisome, with which they share considerable structural and amino acid sequence similarity (Kuhlen et al., 2018; Abrusci et al., 2012; Eichelberg et al., 1994; Johnson et al., 2019). The flagellar export machinery comprises an ATPase complex (FliHIJ) located in the cytoplasm, peripheral to the membrane. Immediately above the ATPase is a nonameric ring formed by the cytoplasmic domain of FlhA (FlhAC), which functions as a subunit docking platform (Bange et al., 2010; Kinoshita et al., 2013; Bryant et al., 2021; Xing et al., 2018). A recent cryo-ET map indicates that the FlhA family have a sea-horse-like structure, in which FlhAc forms the ‘body’ and the FlhA N-terminal region (FlhAN) forms the ‘head’, which is fixed in the plane of the membrane (Butan et al., 2019). FlhAN wraps around the base of a complex formed by FliPQR and the N-terminal sub-domain of FlhB (FlhBN), and together these form the FlhAB-FliPQR export gate that connects the cytoplasm to the central channel in the nascent flagellum, which is contiguous with the extracellular environment (Kuhlen et al., 2018; Abrusci et al., 2012; Eichelberg et al., 1994; Johnson et al., 2019; Bryant et al., 2021; Xing et al., 2018; Butan et al., 2019; Mizuno et al., 2011). FlhBN is connected via a linker (FlhBCN) to the cytoplasmic domain of FlhB (FlhBC), which is thought to sit between the FlhAN and FlhAC rings, where it functions as a docking site for early flagellar subunits (Evans et al., 2013; Kuhlen et al., 2018; Butan et al., 2019).
The ‘early’ flagellar subunits that assemble to form the rod and hook substructures are not chaperoned: instead, the signals for targeting and export are found within the early subunits themselves. We have shown that of one of these signals is a small hydrophobic sequence termed the gate recognition motif (GRM), which is essential for early subunit export (Evans et al., 2013). This motif binds a surface exposed hydrophobic pocket on FlhBc (Evans et al., 2013). Once subunits reach the export machinery, they must be unfolded before they can pass through the narrow channel formed by FliPQR-FlhBN into the central channel of the nascent flagellum, through which the subunits transit until they reach the tip and fold into the structure (Evans et al., 2014b; Kuhlen et al., 2018). Structural studies suggest that FliPQR-FlhBN adopts an energetically favourable closed conformation, possibly to maintain the membrane permeability barrier (Kuhlen et al., 2018; Johnson et al., 2019; Ward et al., 2018; Kuhlen et al., 2020). This suggests that there must be a mechanism to trigger opening of the export gate when subunits dock at cytosolic face of the flagellar export machinery.
Here, we sought to identify new export signals within flagellar rod/hook subunits, using the hook-cap subunit FlgD as a model export substrate. We show that the extreme N-terminus of rod/hook subunits contains a hydrophobic export signal and investigate its functional relationship to the subunit gate recognition motif (GRM).
Results
Identification of a hydrophobic export signal at the N-terminus of FlgD
The N-terminal region of flagellar rod and hook subunits is required for their export (Minamino and Macnab, 1999; Evans et al., 2013). Using the flagellar hook-cap protein FlgD as a model rod/hook subunit, we sought to identify specific export signals within the N-terminus. A screen of ten FlgD variants containing internal five-residue scanning deletions in the first 50 residues (although FlgD∆2–5 is a four-residue deletion, retaining the initial methionine) identified just two variants defective for export into culture supernatant (Figure 1A). Loss of residues 2–5 caused a significant reduction in export, as did deletion of residues 36–40, although to a lesser extent (Figure 1A; Evans et al., 2013). We have shown that FlgD residues 36–40 are the gate recognition motif (GRM) required for transient subunit docking at the FlhBC component of the export gate (Evans et al., 2013). The results suggest that the extreme N-terminus might also be important for interaction with the export machinery.
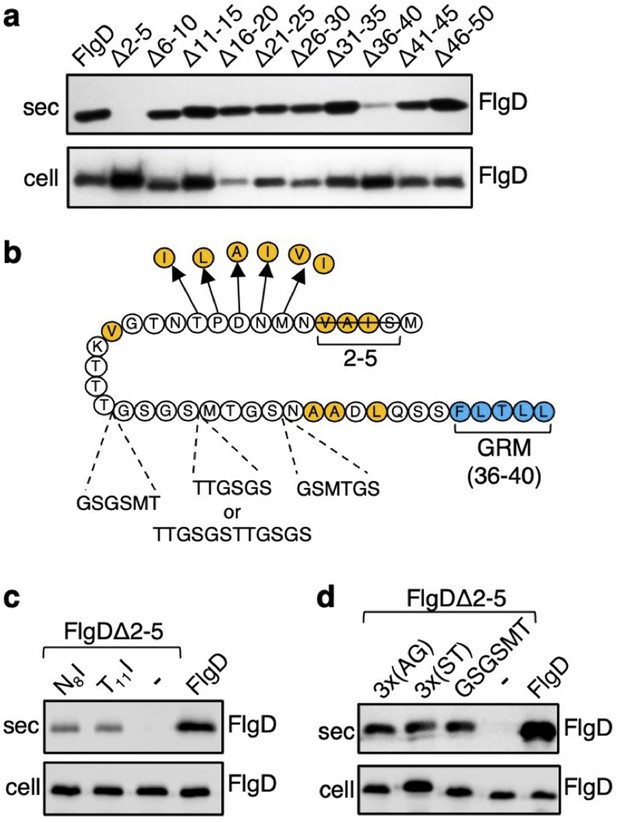
Screening for export-defective FlgD variants.
(a) Whole cell (cell) and supernatant (sec) proteins from late exponential phase cultures of a Salmonella flgD null strain expressing plasmid-encoded wild type FlgD (FlgD) or its variants (Δ2–5, Δ6–10, Δ11–15, Δ16–20, Δ21–25, Δ26–30, Δ31–35, Δ36–40, Δ41–45 or Δ46–50) were separated by SDS (15%)-PAGE and analysed by immunoblotting with anti-FlgD polyclonal antisera. (b) A schematic displaying all intragenic suppressor mutations within amino acids 1–40 of FlgD isolated from the FlgDΔ2–5 variant. Small non-polar residues are highlighted in orange. All suppressor mutations were located between the gate-recognition motif (GRM, blue) and the extreme N-terminus, and can be separated into two classes: insertions or duplications that introduced additional sequence between valine-15 and the gate-recognition motif, or missense mutations that re-introduce small non-polar residues at the N-terminus. All intragenic suppressors isolated from FlgDΔ2–5 are displayed in this figure. (c) Whole cell (cell) and supernatant (sec) proteins from late exponential-phase cultures of Salmonella flgD null strains expressing plasmid-encoded: suppressor mutants isolated from the FlgDΔ2–5 variant (FlgDΔ2–5-N8I or FlgDΔ2–5-T11I), FlgDΔ2–5 variant (-) or wild type FlgD (FlgD) were separated by SDS (15%)-PAGE and analysed by immunoblotting with anti-FlgD polyclonal antisera. (d) Whole cell (cell) and supernatant (sec) proteins from late exponential phase cultures of Salmonella flgD null strains expressing plasmid-encoded wild type FlgD (labelled FlgD), FlgDΔ2–5 (labelled as DΔ2–5) or variants of FlgDΔ2–5 containing between residues 19 and 20 a six-residue insertion of either small non-polar (AGAGAG) residues (labelled as 3 x(AG)), polar (STSTST) residues (labelled as 3 x(ST)), or the sequence from an isolated insertion suppressor mutant (GSGSMT) (labelled as GSGSMT), were separated by SDS (15%)-PAGE and analysed by immunoblotting with anti-FlgD polyclonal antisera.
-
Figure 1—source data 1
Full-length protein western blot of secreted proteins relating to Figure 1A.
- https://cdn.elifesciences.org/articles/66264/elife-66264-fig1-data1-v2.pdf
-
Figure 1—source data 2
Full-length western blot of cellular proteins relating to Figure 1A.
- https://cdn.elifesciences.org/articles/66264/elife-66264-fig1-data2-v2.pdf
-
Figure 1—source data 3
Full-length western blot of cellular and secreted proteins relating to Figure 1C (bottom).
- https://cdn.elifesciences.org/articles/66264/elife-66264-fig1-data3-v2.pdf
-
Figure 1—source data 4
Full-length western blot of cellular proteins relating to Figure 1D (low exposure, bottom).
- https://cdn.elifesciences.org/articles/66264/elife-66264-fig1-data4-v2.pdf
-
Figure 1—source data 5
Full-length western blot of cellular proteins relating to Figure 1D (high exposure, bottom).
- https://cdn.elifesciences.org/articles/66264/elife-66264-fig1-data5-v2.pdf
To gain insight into the putative new signal, we screened for intragenic suppressor mutations that could restore export of the FlgD∆2–5 variant. A Salmonella flgD null strain expressing flgD∆2–5 in trans was inoculated into soft-tryptone agar and incubated until ‘spurs’ of motile cell populations appeared. Sequencing of flgD∆2–5 alleles from these motile populations identified ten different intragenic gain-of-function mutations. These could be separated into two classes (Figure 1B, Figure 1—figure supplement 1).
The first class of motile revertants carried flgD∆2–5 alleles with missense mutations that introduced small non-polar residues at the extreme N-terminus of FlgD∆2–5 (Figure 1B). Deletion of residues 2–5 (2SIAV5) had removed all small non-polar amino acids from the first ten residue region of FlgD, effectively creating a new N-terminus containing a combination of polar, charged or large non-polar residues (Figure 1—figure supplement 2). Analysis of other flagellar rod and hook subunit primary sequences revealed that in every case their native N-terminal regions contain small non-polar residues positioned upstream of the gate recognition motif (GRM residues 36–40; Figure 1—figure supplement 2), indicating that hydrophobicity may be key to the function of the N-terminal export signal. Removal of non-polar residues from the extreme N-terminus of the secreted hook-length control protein, FliK, attenuated its export, indicating that the hydrophobic N-terminal signal is required for export of early subunits (Figure 1—figure supplement 3).
Export assays performed with two representative motile revertant strains carrying flgD∆2–5 variants with gain-of-function point mutations, those encoding FlgD∆2–5-N8I and FlgD∆2–5-T11I, revealed that export of these subunits had recovered to ~50% of the level observed for wild type FlgD (Figure 1C, Figure 1—figure supplements 4–5).
The second class of motile revertants carried flgD∆2–5 alleles that had acquired duplications or insertions introducing at least six additional residues between the FlgD∆2–5 N-terminus and the gate recognition motif (GRM; Figure 1B). It seemed possible that these insertions/duplications might have restored subunit export either by insertion of amino acids that could function as a ‘new’ hydrophobic export signal, or by restoring the position of an existing small hydrophobic residue or sequence relative to the GRM.
To assess these possibilities, we tested whether export of FlgD∆2–5 could be recovered by inserting either polar (19STSTST20) or small non-polar (19AGAGAG20) residues in the FlgD∆2–5 N-terminal region at a position equivalent to one of the suppressing duplications (19GSGSMT20; Figure 1B and D, Figure 1—figure supplements 4–5). We reasoned that if suppression by the additional sequence had been caused by repositioning an existing small hydrophobic amino acid relative to the GRM, then any insertional sequence (polar or non-polar) would restore export, while if suppression had resulted from insertion of a ‘new’ export signal, then either the polar STSTST or non-polar AGAGAG, but not both, could be expected to restore export.
We found that both the engineered FlgD variants (FlgD∆2-5-19AGAGAG20 and FlgD∆2-5-19STSTST20) were exported from a Salmonella flgD null strain as effectively as the gain-of-function mutant FlgD∆2-5-19GSGSMT20 isolated from the suppressor screen (Figure 1D). This suggests that the insertions had repositioned a sequence in the FlgD∆2–5 N-terminus relative to the GRM to overcome the loss of small hydrophobic residues.
The position of the hydrophobic export signal relative to the gate recognition motif is critical for Rod and Hook subunit export
The intragenic suppressor experiments indicated that FlgD export requires a hydrophobic export signal towards the N-terminus and that the position of this hydrophobic signal relative to the previously described GRM is important. Sequence analysis of the gain-of-function FlgD∆2–5 insertion variants revealed that the insertions were all located between the GRM and valine15 (V15; Figure 1B). We reasoned that they repositioned valine15 relative to the GRM, such that it could perform the function of the N-terminal hydrophobic signal lost in FlgD∆2–5. To test this view, we replaced V15 by alanine in the gain-of-function variant FlgD∆2-5-19(GSGSMT)20 and assayed its export in the Salmonella flgD null (Figure 2A, Figure 2—figure supplement 1). Unlike the flgD null strain producing either the parental FlgD∆2-5-19(GSGSMT)20 or wild type FlgD, the flgD null carrying variant FlgD∆2-5-19(GSGSMT)20-V15A was non-motile, reflecting the variant’s failure to export (Figure 2A, Figure 2—figure supplement 1). This suggests that the V15 residue had indeed compensated for the missing N-terminal hydrophobic signal.
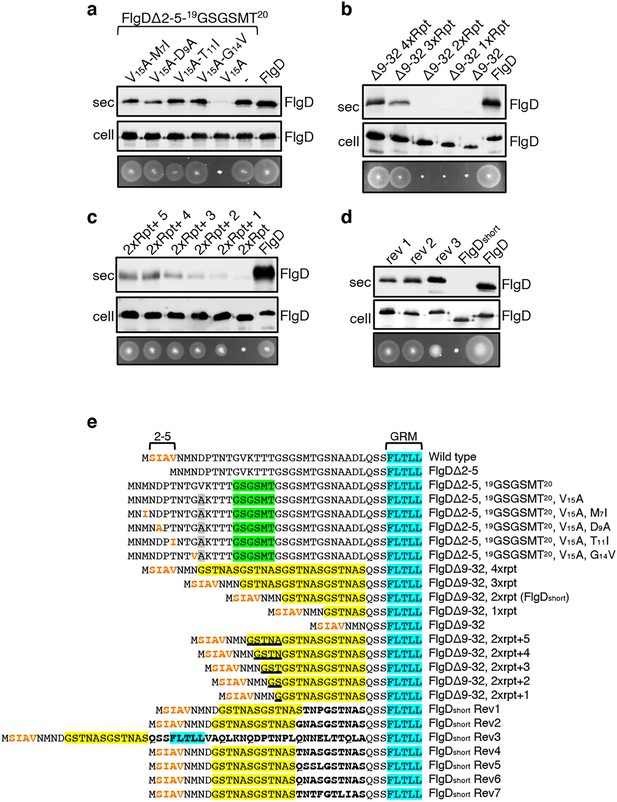
Export of FlgD variants in which the position of the hydrophobic export signal is varied relative to the gate recognition motif (GRM).
Whole cell (cell) and supernatant (sec) proteins from late exponential-phase cultures of a Salmonella flgD null strain expressing plasmid encoded suppressor mutants isolated from the FlgDΔ2-5-19(GSGSMT)20 V15A variant (V15A-M7I, V15A-D9A, V15A-T11I, V15A-G14V), their parent FlgD variant FlgDΔ2-5-19(GSGSMT)20 V15A (labelled as V15A), FlgDΔ2-5-19(GSGSMT)20 (labelled as -) or wild type FlgD (FlgD) were separated by SDS (15%)-PAGE and analysed by immunoblotting with anti-FlgD polyclonal antisera. Swimming motility (bottom panel; 0.25% soft tryptone agar) of the same strains were carried out at 37 °C for 4–6 hours. (b). Whole cell (cell) and supernatant (sec) proteins from late exponential-phase cultures of a Salmonella flgD null strain expressing plasmid-encoded wild type FlgD (labelled as FlgD), FlgDΔ9–32 or its variants in which residues 9–32 were replaced by between one and four six-residue repeats of Gly-Ser-Thr-Asn-Ala-Ser (GSTNAS): (Δ9–32 4xRpt, Δ9–32 3xRpt, Δ9–32 2xRpt or Δ9–32 1xRpt) were separated by SDS (15%)-PAGE and analysed by immunoblotting with anti-FlgD polyclonal antisera. Swimming motility (bottom panel; 0.25% soft tryptone agar) of the same strains were carried out at 37 °C for 4–6 hours. (c). Whole cell (cell) and supernatant (sec) proteins from late exponential-phase cultures of a Salmonella flgD null strain expressing plasmid-encoded wild type FlgD (labelled as FlgD), a FlgD variant in which residues 9–32 were replaced by two repeats of a six-residue sequence Gly-Ser-Thr-Asn-Ala-Ser (labelled as 2xRpt) or its variants containing between one and five additional residues inserted directly after the two repeats (labelled as 2xRpt + 1, 2xRpt + 2, 2xRpt + 3, 2xRpt + 4 or 2xRpt + 5) were separated by SDS (15%)-PAGE and analysed by immunoblotting with anti-FlgD polyclonal antisera. Swimming motility (bottom panel; 0.25% soft tryptone agar) of the same strains were carried out at 37 °C for 4–6 hr. (d). Whole cell (cell) and supernatant (sec) proteins from late exponential-phase cultures of a Salmonella flgD null strain expressing plasmid-encoded wild type FlgD (labelled as FlgD), a FlgD variant in which residues 9–32 were replaced by two repeats of a six-residue sequence Gly-Ser-Thr-Asn-Ala-Ser (labelled as FlgDshort) or suppressor mutants isolated from this strain (labelled as rev1, rev2 or rev3) were separated by SDS (15%)-PAGE and analysed by immunoblotting with anti-FlgD polyclonal antisera. Swimming motility (bottom panel; 0.25% soft tryptone agar) of the same strains were carried out at 37 °C for 4–6 hr. (e). N-terminal sequences of wild type FlgD and its variants aligned to their gate-recognition motif (GRM; blue). The following sequence features or residues are displayed: The N-terminal hydrophobic signal (residue 2–5; orange), the Gly-Ser-Gly-Ser-Met-Thr (GSGSMT) insertion (green) isolated from the FlgDΔ2–5 suppressor screen, the valine-15 to alanine mutation (grey), small non-polar mutations (M7I, D9A, T11I, G14V; orange) isolated from the FlgDΔ2–5, 19GSGSMT20 suppressor screen, FlgDΔ9–32 and its variants in which residues 9–32 are replaced with one, two, three or four repeats of a six-residue sequence Gly-Ser-Thr-Asn-Ala-Ser (GSTNAS; yellow), FlgDΔ9–32, 2xrpt (hereafter termed FlgDshort) containing five, four, three, two, or one additional residues (underlined) inserted between the GRM and N-terminal hydrophobic signal, and suppressor mutants (Rev 1–7) isolated from FlgDshort that introduced additional residues (bold) between the N-terminal hydrophobic signal (orange) and the gate-recognition motif (blue).
-
Figure 2—source data 1
Full-length protein western blot of cellular and secreted proteins relating to Figure 2A.
- https://cdn.elifesciences.org/articles/66264/elife-66264-fig2-data1-v2.pdf
-
Figure 2—source data 2
Full-length protein western blot of cellular and secreted proteins relating to Figure 2B (bottom).
- https://cdn.elifesciences.org/articles/66264/elife-66264-fig2-data2-v2.pdf
-
Figure 2—source data 3
Full-length protein western blot of cellular proteins relating to Figure 2C (bottom).
- https://cdn.elifesciences.org/articles/66264/elife-66264-fig2-data3-v2.pdf
-
Figure 2—source data 4
Full-length protein western blot of secreted proteins relating to Figure 2C (bottom).
- https://cdn.elifesciences.org/articles/66264/elife-66264-fig2-data4-v2.pdf
-
Figure 2—source data 5
Full-length protein western blot of cellular proteins relating to Figure 2D (bottom).
- https://cdn.elifesciences.org/articles/66264/elife-66264-fig2-data5-v2.pdf
-
Figure 2—source data 6
Full-length protein western blot of secreted proteins relating to Figure 2D (bottom, left).
- https://cdn.elifesciences.org/articles/66264/elife-66264-fig2-data6-v2.pdf
By screening for intragenic suppressors of the motility defect associated with FlgD∆2-5-19(GSGSMT)20-V15A, four gain-of-function missense mutations were identified, M7I, D9A, T11I and G14V. All these had introduced small hydrophobic residues, all positioned at least 27 residues upstream of the GRM. These FlgD∆2-5-19(GSGSMT)20-V15A gain-of-function variants restored motility to the Salmonella flgD null strain and were exported at levels similar to wildtype FlgD and FlgD∆2-5-19(GSGSMT)20 (Figure 2A, Figure 2—figure supplement 1). These data confirm the importance of small non-polar residues positioned upstream of the GRM.
Our results so far had indicated that the position of the FlgD N-terminal hydrophobic export signal relative to the GRM was critical and suggested that, for export to occur efficiently, at least 26 residues must separate the hydrophobic signal and the GRM (Figure 1C, Figure 1—figure supplement 4). In the primary sequences of all Salmonella flagellar rod/hook subunits the GRM is positioned ≥30 amino acids downstream of the subunit N-terminus (Figure 1—figure supplement 4), suggesting that separation of the two signals by a minimum number of residues might be a common feature among early flagellar subunits. To test this, a suite of engineered flgD alleles was constructed that encoded FlgD variants in which wildtype residues 9–32 were replaced with between one and four repeats of the six amino acid sequence Gly-Ser-Thr-Asn-Ala-Ser (GSTNAS). Swimming motility and export assays revealed that the minimum number of inserted GSTNAS repeats that could support efficient FlgD export was three, equivalent to separation of the hydrophobic N-terminal signal and the GRM by 24 residues (Figure 2B). Below this threshold, FlgD export and swimming motility were strongly attenuated (Figure 2B). A further set of recombinant flgD alleles was constructed, which encoded FlgD∆9–32 variants carrying two GSTNAS repeats (hereafter termed FlgDshort) directly followed by between one and five additional residues (Figure 2E). Motility and FlgD export increased incrementally with the addition of each amino acid (Figure 2C, Figure 2—figure supplement 1). The data indicate that a low level of FlgD export is supported when the hydrophobic N-terminal signal (2SIAV5) and the GRM (36FLTLL40) are separated by 19 residues, with export efficiency and swimming motility increasing as separation of the export signals approaches an optimal 24 residues.
To further establish the requirement for a minimum number of residues between the hydrophobic N-terminal signal and the GRM, we screened for intragenic suppressor mutations that could restore swimming motility in a flgD null strain producing FlgDshort. Sequencing of flgDshort alleles from motile revertant strains identified seven gain-of-function mutations that introduced additional residues between the hydrophobic N-terminal signal and the GRM (Figure 2—figure supplement 1). Swimming motility and FlgD export was assessed for three flgD null strains expressing representative flgDshort gain-of-function variants and all showed increased FlgD subunit export and swimming motility compared to the flgD null expressing flgDshort (Figure 2D, Figure 2—figure supplement 1). The data confirm that the position of the hydrophobic N-terminal signal relative to the GRM is critical for efficient FlgD subunit export.
To establish that this is a general requirement for the export of other rod and hook subunits, engineered alleles of flgE (hook) and flgG (rod) were constructed that encoded variants in which FlgE residues 9–32 or FlgG residues 11–35 were either deleted (FlgEshort and FlgGshort) or replaced with four repeats of the sequence GSTNAS (Figure 3). As had been observed for FlgDshort, export of the FlgEshort and FlgGshort variants was severely attenuated compared to wild-type FlgE and FlgG (Figure 3, Figure 2—figure supplement 2). Furthermore, insertion of four GSTNAS repeats into FlgEshort and FlgGshort recovered subunit export to wild-type levels, indicating that the minimum separation of the hydrophobic N-terminal signal and the GRM is a feature throughout rod and hook subunits (Figure 3).
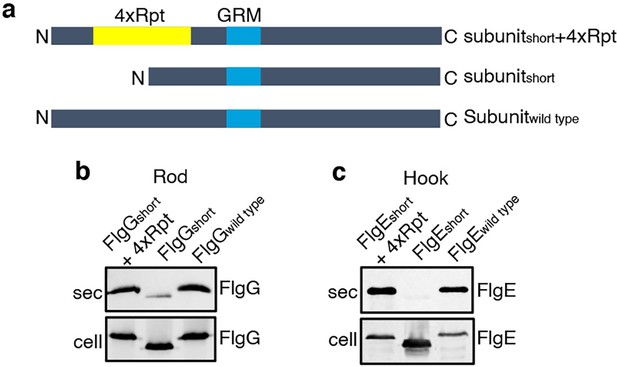
Effect of the relative position of the N-terminus and GRM on the export of other rod and hook subunits.
(a) Schematic representation of a wild-type subunit (labelled as subunitwild type), a subunit containing a deletion of sequence from between the N-terminus and GRM (labelled as subunitshort) and a subunit in which the deleted sequence was replaced by four repeats of a six-residue sequence Gly-Ser-Thr-Asn-Ala-Ser (yellow, labelled as subunitshort +4 Rpt). (b). Whole cell (cell) and supernatant (sec) proteins from late exponential-phase cultures of a Salmonella flgE null strain expressing plasmid-encoded wild type FlgG (labelled as FlgGwild type), a FlgG variant in which residues 11–35 were deleted (labelled as FlgGshort) or a FlgG variant in which residues 11–35 were replaced by four repeats of a six-residue sequence Gly-Ser-Thr-Asn-Ala-Ser (labelled as FlgGshort + 4 Rpt). All FlgG variants were engineered to contain an internal 3xFLAG tag for immunodetection. Proteins were separated by SDS (15%)-PAGE and analysed by immunoblotting with anti-FLAG monoclonal antisera. (c). Whole cell (cell) and supernatant (sec) proteins from late exponential-phase cultures of a Salmonella flgD null strain expressing plasmid-encoded wild type FlgE (labelled as FlgEwild type), a FlgE variant in which residues 9–32 were deleted (labelled as FlgEshort) or a FlgE variant in which residues 9–32 were replaced by four repeats of a six-residue sequence Gly-Ser-Thr-Asn-Ala-Ser (labelled as FlgEshort + 4 Rpt). All FlgE variants were engineered to contain an internal 3xFLAG tag for immunodetection. Proteins were separated by SDS (15%)-PAGE and analysed by immunoblotting with anti-FLAG monoclonal antisera.
-
Figure 3—source data 1
Full-length protein western blot of cellular and secreted proteins relating to Figure 3B (low exposure).
- https://cdn.elifesciences.org/articles/66264/elife-66264-fig3-data1-v2.pdf
-
Figure 3—source data 2
Full-length protein western blot of cellular and secreted proteins relating to Figure 3B (high exposure).
- https://cdn.elifesciences.org/articles/66264/elife-66264-fig3-data2-v2.pdf
-
Figure 3—source data 3
Full-length protein western blot of secreted proteins relating to Figure 3C (low exposure, top).
- https://cdn.elifesciences.org/articles/66264/elife-66264-fig3-data3-v2.pdf
-
Figure 3—source data 4
Full-length protein western blot of cellular proteins relating to Figure 3C (high exposure, bottom).
- https://cdn.elifesciences.org/articles/66264/elife-66264-fig3-data4-v2.pdf
Sequential engagement of the subunit GRM and hydrophobic N-terminal export signal by the flagellar export machinery
Having identified a new hydrophobic N-terminal export signal and established that its position relative to the GRM was critical, we next wanted to determine the order in which the signals were recognised/engaged by the export machinery. The signals might be recognised simultaneously, with both being required for initial entry of rod/hook subunits into the export pathway. Alternatively, they might be recognised sequentially. If this were the case, then a subunit variant that possessed the ‘first’ signal but was deleted for the ‘second’ signal might enter the export pathway but fail to progress, becoming stalled at a specific step to block the pathway and prevent export of wild type subunits. To test if FlgD∆2–5 or FlgD∆GRM stalled in the export pathway, recombinant expression vectors encoding these variants or wild type FlgD were introduced into a Salmonella ∆recA strain that is wild type for flagellar export (Figure 4). We could then assess whether the variant FlgD constructs could interfere in trans with the wildtype flagellar export. To do this, we assessed the export of an early flagellar substrate, FliK, which controls the length of the flagellar hook and of the late export substrate, FlgK and FlgL, which together form a junction connecting the flagellar filament to the hook. We saw that FlgD∆2–5 inhibited motility and export of the FliK, FlgK and FlgL flagellar subunits, whereas FlgD∆GRM did not (Figure 4B and C). The data indicate that FlgD∆2–5 enters the flagellar export pathway and stalls at a critical point, blocking export. In contrast, FlgD∆GRM does not stall or block export.
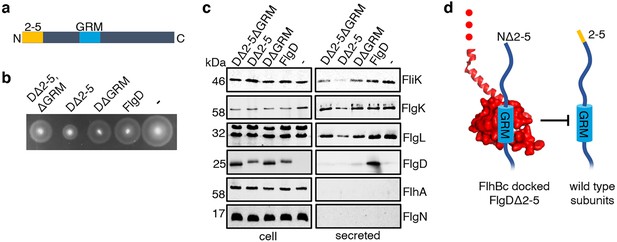
Effect on subunit export of overexpressed FlgDΔ2–5 and variants.
(a) Schematic representation of a FlgD subunit containing a N-terminal hydrophobic signal (orange, labelled as 2–5) and a gate-recognition motif (blue, labelled as GRM). (b) Swimming motility of a Salmonella ΔrecA strain expressing plasmid-encoded wild type FlgD (FlgD), its variants (DΔ2-5ΔGRM, DΔ2–5 or DΔGRM) or empty pTrc99a vector (-). Motility was assessed in 0.25% soft-tryptone agar containing 100 μg/ml ampicillin and 100 μM IPTG and incubated for 4–6 hr at 37 °C. (c) Whole cell (cell) and secreted proteins (secreted) from late-exponential-phase cultures were separated by SDS (15%)-PAGE and analysed by immunoblotting with anti-FliK (hook ruler subunit), anti-FlgK and anti-FlgL (hook-filament junction subunits), anti-FlgD hook cap subunit, anti-FlhA (component of the export machinery) and anti-FlgN (export chaperone for FlgK and FlgL) polyclonal antisera. Apparent molecular weights are in kilodaltons (kDa).(d). A model depicting a FlgDΔ2–5 subunit (left) docked via its gate-recognition motif (GRM, blue) at the subunit binding pocket on FlhBC (PDB: 3B0Z Kuhlen et al., 2020, red), preventing wild type subunits (right) from docking at FlhBC.
-
Figure 4—source data 1
Full-length protein western blot of cellular and secreted proteins relating to Figure 4C (anti-FliK, bottom).
- https://cdn.elifesciences.org/articles/66264/elife-66264-fig4-data1-v2.pdf
-
Figure 4—source data 2
Full-length protein western blot of cellular and secreted proteins relating to Figure 4C (anti-FlgK, top).
- https://cdn.elifesciences.org/articles/66264/elife-66264-fig4-data2-v2.pdf
-
Figure 4—source data 3
Full-length protein western blot of cellular and secreted proteins relating to Figure 4C (anti-FlgL).
- https://cdn.elifesciences.org/articles/66264/elife-66264-fig4-data3-v2.pdf
-
Figure 4—source data 4
Full-length protein western blot of cellular proteins relating to Figure 4C (anti-FlgD, top right).
- https://cdn.elifesciences.org/articles/66264/elife-66264-fig4-data4-v2.pdf
-
Figure 4—source data 5
Full-length protein western blot of secreted proteins relating to Figure 4C (anti-FlgD, bottom).
- https://cdn.elifesciences.org/articles/66264/elife-66264-fig4-data5-v2.pdf
-
Figure 4—source data 6
Full-length protein western blot of cellular and secreted proteins relating to Figure 4C (anti-FlhA (top) and anti-FlgN (bottom)).
- https://cdn.elifesciences.org/articles/66264/elife-66264-fig4-data6-v2.pdf
To determine whether FlgD∆2–5 stalls at a point before or after subunit docking at the FlhBC component of the flagellar export gate via the GRM, a recombinant vector encoding a FlgD variant in which both export signals were deleted (FlgD∆2–5∆GRM) was constructed. If loss of the hydrophobic N-terminal signal had caused subunits to stall after docking at FlhBC, then additional deletion of the subunit GRM would relieve this block. Motility and subunit export assays revealed that the Salmonella ∆recA strain producing FlgD∆2–5∆GRM displayed swimming motility and levels of FliK and FlgK subunit export similar to cells producing FlgD∆GRM (Figure 4B and C). The data suggest that FlgD∆2–5 stalls after docking at the FlhBC export gate, preventing docking of other early subunits.
It seemed possible that subunit docking via the GRM to the FlhBC export gate might position the hydrophobic N-terminal signal in close proximity to its recognition site on the export machinery. If this were the case, ‘short’ subunit variants containing deletions that decreased the number of residues between the hydrophobic N-terminal signal and the GRM might also stall at FlhBC, and this stalling might be relieved by additional deletion of the GRM. To test this, recombinant expression vectors encoding ‘short’ subunit variants (FlgEshort or FlgDshort), ‘short’ subunit variants additionally deleted for the GRM (FlgEshort∆GRM or FlgDshort∆GRM) or wild type FlgE or FlgD were introduced into a Salmonella ∆recA strain (Figure 5). Compared to the wild-type subunits expressed in trans, the ‘short’ subunits inhibited swimming motility and the export of other flagellar subunits (FliD, FliK, FlgK), whereas FlgEshort∆GRM and FlgDshort∆GRM did not (Figure 5). Taken together with the data presented in Figure 4, the results indicate that the subunit GRM and the hydrophobic N-terminal signal are recognised sequentially, with subunits first docking at FlhBC via the GRM, which positions the hydrophobic N-terminal signal for subsequent interactions with the export machinery (Figure 4D).
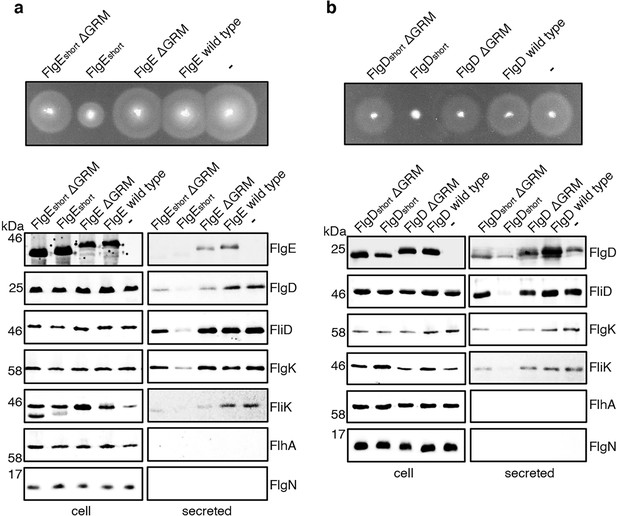
Effect on subunit export of overexpressed FlgEshort, FlgDshort and variants.
(a) Swimming motility of a Salmonella ΔrecA strain expressing plasmid-encoded wild type FlgE (labelled as FlgE wild type), a FlgE variant in which residues 9–32 were deleted (labelled as FlgEshort), a FlgE variant in which residues 9–32 and residues 39–43 (corresponding to the gate-recognition motif) were deleted (labelled as FlgEshortΔGRM), a FlgE variant in which residues 39–43 were deleted (labelled as FlgEΔGRM) or empty pTrc99a vector (labelled as -). All FlgE variants were engineered to contain an internal 3xFLAG tag for immunodetection. Motility was assessed in 0.25% soft-tryptone agar containing 100 μg/ml ampicillin and 100 μM IPTG and incubated for 4–6 hr at 37 °C (top panel). Whole cell (cell) and secreted proteins (secreted) from late-exponential-phase cultures were separated by SDS (15%)-PAGE and analysed by immunoblotting with anti-FLAG monoclonal antisera (to detect the flag tagged hook subunit FlgE) or anti-FlgD (hook cap subunit), anti-FliD (filament cap subunit), anti-FlgK (hook-filament junction subunit), anti-FliK (hook-ruler subunit), anti-FlhA (component of the export machinery) and anti-FlgN (chaperone for FlgK and FlgL) polyclonal antisera (bottom). Apparent molecular weights are in kilodaltons (kDa).(b) Swimming motility of a Salmonella ΔrecA strain expressing plasmid-encoded wild type FlgD (labelled as FlgD wild type), a FlgD variant in which residues 9–32 were replaced with two repeats of the six amino acid sequence Gly-Ser-Thr-Asn-Ala-Ser (labelled as FlgDshort), a FlgD variant in which residues 9–32 were replaced with two repeats of the six amino acid sequence Gly-Ser-Thr-Asn-Ala-Ser and residues 36–40 were deleted (labelled FlgDshortΔGRM), a FlgD variant in which residues 36–40 were deleted (labelled as FlgDΔGRM) or empty pTrc99a vector (labelled as -). Motility was assessed in 0.25% soft-tryptone agar containing 100 μg/ml ampicillin and 100 μM IPTG and incubated for 4–6 hours at 37 °C (top panel). Whole cell (cell) and secreted proteins (secreted) from late-exponential-phase cultures were separated by SDS (15%)-PAGE and analysed by immunoblotting with anti-FlgD (hook cap subunit), anti-FliD (filament cap subunit), anti-FlgK (hook-filament junction subunit), anti-FliK (hook ruler subunit), anti-FlhA (component of export machinery), and anti-FlgN (chaperone for FlgK and FlgL) polyclonal antisera (bottom). Apparent molecular weights are in kilodaltons (kDa).
-
Figure 5—source data 1
Full-length protein western blot of cellular proteins relating to Figure 4A (low contrast, anti-FLAG).
- https://cdn.elifesciences.org/articles/66264/elife-66264-fig5-data1-v2.pdf
-
Figure 5—source data 2
Full-length protein western blot of secreted proteins relating to Figure 4A (high contrast, anti-FLAG).
- https://cdn.elifesciences.org/articles/66264/elife-66264-fig5-data2-v2.pdf
-
Figure 5—source data 3
Full-length protein western blot of cellular and secreted proteins relating to Figure 4A (anti-FliD (top) and anti-FlgD (bottom)).
- https://cdn.elifesciences.org/articles/66264/elife-66264-fig5-data3-v2.pdf
-
Figure 5—source data 4
Full-length protein western blot of cellular and secreted proteins relating to Figure 4A (anti-FlgK).
- https://cdn.elifesciences.org/articles/66264/elife-66264-fig5-data4-v2.pdf
-
Figure 5—source data 5
Full-length protein western blot of cellular proteins relating to Figure 4A (anti-FliK, left).
- https://cdn.elifesciences.org/articles/66264/elife-66264-fig5-data5-v2.pdf
-
Figure 5—source data 6
Full-length protein western blot of secreted proteins relating to Figure 4A (anti-FliK, bottom right).
- https://cdn.elifesciences.org/articles/66264/elife-66264-fig5-data6-v2.pdf
-
Figure 5—source data 7
Full-length protein western blot of cellular and secreted proteins relating to Figure 4A (anti-FlhA, top).
- https://cdn.elifesciences.org/articles/66264/elife-66264-fig5-data7-v2.pdf
-
Figure 5—source data 8
Full-length protein western blot of cellular and secreted proteins relating to Figure 4A (anti-FlgN, bottom).
- https://cdn.elifesciences.org/articles/66264/elife-66264-fig5-data8-v2.pdf
-
Figure 5—source data 9
Full-length protein western blot of cellular proteins relating to Figure 4B (anti-FlgD, left).
- https://cdn.elifesciences.org/articles/66264/elife-66264-fig5-data9-v2.pdf
-
Figure 5—source data 10
Full-length protein western blot of secreted proteins relating to Figure 4B (anti-FlgD, bottom right).
- https://cdn.elifesciences.org/articles/66264/elife-66264-fig5-data10-v2.pdf
-
Figure 5—source data 11
Full-length protein western blot of cellular and secreted proteins relating to Figure 4B (anti-FliD).
- https://cdn.elifesciences.org/articles/66264/elife-66264-fig5-data11-v2.pdf
-
Figure 5—source data 12
Full-length protein western blot of cellular and secreted proteins relating to Figure 4B (anti-FlgK, top).
- https://cdn.elifesciences.org/articles/66264/elife-66264-fig5-data12-v2.pdf
-
Figure 5—source data 13
Full-length protein western blot of cellular and secreted proteins relating to Figure 4B (anti-FliK).
- https://cdn.elifesciences.org/articles/66264/elife-66264-fig5-data13-v2.pdf
-
Figure 5—source data 14
Full-length protein western blot of cellular and secreted proteins relating to Figure 4B (anti-FlhA (top) and anti-FlgN (bottom)).
- https://cdn.elifesciences.org/articles/66264/elife-66264-fig5-data14-v2.pdf
Mutations that promote opening of the export gate partially compensate for incorrect positioning of the N-terminal export signal
The accruing data indicated that subunit docking at FlhBC might correctly position the hydrophobic N-terminal signal for recognition by the export machinery. To model the position of FlhBC relative to other components of the export machinery, we docked the structures of FlhBC and the FliPQR-FlhBN export gate into the tomographic reconstruction of the Salmonella SPI-1 type III secretion system (Figure 6B; Meshcheryakov et al., 2013). The model indicated that FliPQR-FlhBN and the subunit docking site on FlhBC are separated by a minimum distance of ~78 Å, and that FlhBC is positioned no more than ~22–45 Å from FlhA (Figure 6A; Butan et al., 2019). Taking FlgD as a model early flagellar subunit, the distance between the FlgD N-terminal hydrophobic signal and the GRM was found to be in the range of ~45 Å (α-helix) to ~105 Å (unfolded contour length), depending on the predicted structure adopted by the subunit N-terminus (Figure 6A). Based on these estimates, it seemed feasible that the hydrophobic N-terminal signal of a subunit docked at FlhBC could contact either FlhA or the FliPQR-FlhBN complex, and that this interaction might trigger opening of the export gate (Kuhlen et al., 2018; Meshcheryakov et al., 2013; Bryant and Fraser, 2021; Vonderviszt et al., 1992). If this were true, mutations that promote the open conformation of the export gate might compensate for the incorrect positioning of the hydrophobic N-terminal export signal in ‘short’ rod/hook subunits. One such export gate mutation, FliP-M210A, has been shown to increase ion conductance across the bacterial inner membrane, indicating that this gate variant fails to close efficiently (Kuhlen et al., 2020).
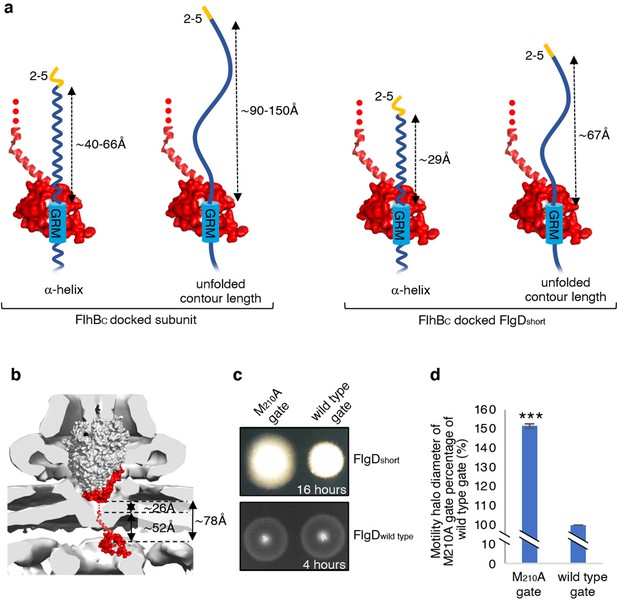
Suppression of the FlgDshort motility defect by mutations in FliP.
(a) A model depicting subunits docked via their gate-recognition motif (GRM, blue) at the subunit binding pocket on FlhBC (PDB: 3B0Z Meshcheryakov et al., 2013, red) with N-termini of early flagellar subunits adopting either an α-helical conformation separating the N-terminal hydrophobic signal (2–5, orange) and gate-recognition motif (GRM, blue) by ~40–60 ångstrom (where each amino acid is on average separated by ~1.5 Å, left) or an unfolded conformation where the unfolded contour length separating the N-terminal hydrophobic signal (2-5) and gate-recognition motif (GRM) is ~90–150 ångstrom (where each amino acid is on average separated by ~3.5 Å, middle left). Values corresponding to the distance separating the N-terminal hydrophobic signal (2–5, orange) and gate-recognition motif (GRM, blue) of a FlgD subunit variant in which residues 9–32 are replaced with two repeats of the six amino acid sequence Gly-Ser-Thr-Asn-Ala-Ser (FlgDshort) indicate that the N-terminal hydrophobic signal (2–5, orange) and gate-recognition motif (GRM, blue) are separated by ~29 ångstrom (α-helical conformation, middle right) or ~67 ångstrom (unfolded contour length, right). (b) Placement of the crystal structure of FlhBc (PDB:3BOZ Kuhlen et al., 2020; red) and the cryo-EM structure of FliPQR-FlhB (PDB:6S3L Ward et al., 2018) in a tomographic reconstruction of the Salmonella SPI-1 injectisome (EMD-8544 [60]; grey). The minimum distance between the subunit gate-recognition motif binding site on FlhBC (grey) to FlhBN (defined as Salmonella FlhB residue 211 [61]; ~ 78 Å) was estimated by combining: the value corresponding to the distance between the subunit binding pocket on FlhBC (Evans et al., 2013) (grey) and the N-terminal visible residue (D229) in the FlhBC structure (PDB:3BOZ Kuhlen et al., 2020; ~ 52 Å) with the value corresponding to the minimum distance between FlhB residues 211 and 228 (based on a linear α-helical conformation; ~ 26 Å). (c) Swimming motility of recombinant Salmonella flgD null strains producing a chromosomally-encoded FliP-M210A variant (M210A gate, left) or wild type FliP (wild-type gate, right). Wildtype FliP and FliP-M210A were engineered to contain an internal HA tag positioned between residue 21 and 22 to allow immunodetection of FliP. Both strains produced either a pTrc99a plasmid-encoded FlgD subunit variant in which residues 9–32 were replaced with two repeats of the six amino acid sequence Gly-Ser-Thr-Asn-Ala-Ser (FlgDshort; top panel) or a pTrc99a plasmid-encoded wild-type FlgD subunit (FlgDwild type; bottom panel). Motility was assessed in 0.25% soft-tryptone agar containing 100 μg/ml ampicillin and 50 μM IPTG and incubated for 16 hr (top panel) or 4–6 hr at 37 °C (bottom panel). (d). The mean motility halo diameter of recombinant Salmonella flgD null strains producing a chromosomally-encoded FliP-M210A variant (M210A gate, left) or wild-type FliP (wild-type gate, right). Wild-type FliP and FliP-M210A were engineered to contain an internal HA tag positioned between residue 21 and 22 to allow immunodetection of FliP. Both strains produced a pTrc99a plasmid-encoded FlgD subunit variant in which residues 9–32 were replaced with two repeats of the six amino acid sequence Gly-Ser-Thr-Asn-Ala-Ser (FlgDshort). Error bars represent the standard error of the mean calculated from at least three biological replicates. *** indicates a p-value < 0.001.
To test whether the FliP-M210A variant gate could promote export of ‘short’ subunits, in which the distance between the hydrophobic N-terminal signal and the GRM was reduced, a recombinant expression vector encoding FlgDshort was introduced into Salmonella flgD null strains in which the fliP gene had been replaced with recombinant genes encoding either a functional FliP variant with an internal HA-tag (designated wild-type gate) or the equivalent HA-tagged FliP-M210A variant (designated M210A gate; Figure 6C, Figure 6—figure supplement 1). The swimming motility of these strains was found to be consistently stronger in the strain producing the M210A gate compared to the strain with the wild-type gate, with the motility halo of the fliP-M210A-∆flgD strain expressing FlgDshort having a 50% greater diameter than that of the wild type fliP-∆flgD strain expressing FlgDshort (Figure 6D, Figure 6—figure supplement 1). This increase in motility indicated that the defect caused by incorrect positioning of the hydrophobic N-terminal signal relative to the GRM in FlgDshort could indeed be partially compensated by promoting the gate open conformation.
Discussion
T3SS substrates contain N-terminal export signals, but these have not been fully defined and how they promote subunit export remains unclear. Here, we characterised a new hydrophobic N-terminal export signal in early flagellar rod/hook subunits and showed that the position of this signal relative to the known subunit gate recognition motif (GRM) is key to subunit export.
Loss of the hydrophobic N-terminal signal in the hook cap subunit FlgD had a stronger negative effect on subunit export than deletion of the GRM that enables subunit docking at FlhBC, suggesting that the hydrophobic N-terminal signal may be required to trigger an essential export step. A suppressor screen showed that the export defect caused by deleting the hydrophobic N-terminal signal could be overcome by mutations that either reintroduced small non-polar amino acids positioned 3–7 residues from the subunit N-terminus (e.g. M7I), or introduced additional residues between V15 and the GRM. In such ‘gain of function’ strains containing insertions, changing V15 to alanine abolished subunit export, which was rescued by re-introduction of small non-polar residues close to the N-terminus. These data point to an essential export function for small non-polar residues close to the N-terminus of rod/hook subunits.
It was fortuitous that we chose FlgD as the model for early flagellar subunit. All early subunits contain small hydrophobic residues close to the N-terminus, but FlgD is unique in that only four (I3, A4, V5 and V15) of its first 25 residues are small and non-polar (Figure 1—figure supplement 1). Indeed, there are only three other small hydrophobic residues between the FlgD N-terminus and the GRM (Figure 1, Figure 1—figure supplement 1, Figure 6—figure supplement 2). While deletion of residues 2–5 in FlgD removes the critical hydrophobic N-terminal signal, similar deletions in the N-terminal regions of other rod/hook subunits reposition existing small non-polar residues close to the N-terminus (Figure 1—figure supplement 1, Figure 6—figure supplement 2). This is perhaps why previous deletion studies in early flagellar subunits have failed to identify the hydrophobic N-terminal signal (Evans et al., 2013; Hirano et al., 2005; Minamino et al., 1999).
The finding that subunits lacking the hydrophobic N-terminal export signal, but not the GRM, stalled during export suggested that these two signals were recognised by the flagellar export machinery in a specific order. Mutant variants of other flagellar export substrates or export components have been observed to block the export pathway. For example, a FlgN chaperone variant lacking the C-terminal 20 residues stalls at the FliI ATPase (Thomas et al., 2004), while a GST-tagged FliJ binds FlhA but is unable to associate correctly with FliI so blocking wild type FliJ interaction with FlhA (Ibuki et al., 2012). These attenuations can be reversed by further mutations that disrupt the stalling interactions. This was also observed for FlgD∆2–5. Loss of the hydrophobic N-terminal signal resulted in a dominant-negative effect on motility and flagellar export, but this was abolished by subsequent deletion of the GRM. This indicates that FlgD∆2–5 stalls in the export pathway at FlhBC, blocking the binding site for early flagellar subunits. These data are consistent with sequential recognition of the two export signals: the GRM first docking subunits at FlhBC, and positioning the hydrophobic N-terminal signal to trigger the next export step.
The position of the subunit hydrophobic N-terminal export signal relative to the GRM appears critical for export. Engineering of flgD to encode variants in which the region between the N-terminus and the GRM was replaced with polypeptide sequences of varying lengths showed that these signals must be separated by a minimum of 19 residues for detectable export, with substantial export requiring separation by 30 residues (Figure 3). When subunits dock at FlhBC, which is likely situated within or just below the plane of the inner membrane, the hydrophobic N-terminal signal is positioned close to the FlhAB-FliPQR export gate (Figure 6B). Subunits in which the GRM and N-terminal signal are brought closer together stall at FlhBC, suggesting that the hydrophobic N-terminal signal is unable to contact its recognition site on the export machinery (Figures 5, 6A and B). In all flagellar rod/hook subunits, the GRM is positioned at least 30 residues from the N-terminus (Figure 1—figure supplement 1, Figure 6—figure supplement 2). The physical distance between the two signals will depend on the structure adopted by the subunit N-terminus (Figure 6A). The N-terminal region of flagellar subunits is often unstructured in solution (Kuwajima et al., 1989; Minamino and Macnab, 1999; Kornacker and Newton, 1994; Evans et al., 2013; Végh et al., 2006; Vonderviszt et al., 1992), and such disorder may be an intrinsic feature of flagellar export signals (Kuwajima et al., 1989; Minamino and Macnab, 1999; Kornacker and Newton, 1994; Evans et al., 2013; Végh et al., 2006; Weber-Sparenberg et al., 2006; Aizawa et al., 1990), as they are typical in other bacterial export N-terminal substrate signals such as those of the Sec and Tat systems (Tsirigotaki et al., 2017; Palmer and Berks, 2012). Unstructured signals may facilitate multiple interactions with different binding partners during export, and in the case of export systems that transport unfolded proteins they may aid initial entry of substrates into narrow export channels (Tsirigotaki et al., 2017).
As yet, nothing is known about the structure of the subunit N-terminal domain upon interaction with the flagellar export machinery. Signal peptides in TAT pathway substrates switch between disordered and α-helical conformations depending on the hydrophobicity of the environment (San Miguel et al., 2003). It therefore seems likely that local environments along the flagellar export pathway will influence the conformation of subunit export signals (Kuhlen et al., 2018; Erhardt et al., 2017). Interestingly, FlgD variants that contain additional sequence that position the N-terminal export signal and GRM further apart far above the required threshold did not impede export, which argues that the sequence between both export signals is unfolded (Figure 2).
If the region between the hydrophobic N-terminal signal and the GRM is unstructured and extended, this would correspond to a polypeptide contour length of approximately 72–105 Å (where the length of one amino acid is ~3.6 Å). If the same region were to fold as an α-helix, its length would be approximately 30–36 Å (where one amino acid rises every ~1.5 Å). Placement of the AlphaFold predicted structure of full length FlhB into a tomogram of the T3SS suggests that FlhBC is positioned below the plane of the inner membrane but above the nonameric ring of FlhAC (Figure 6—figure supplement 3). Without further structural information on subunit interactions with the flagellar export machinery and the precise position of FlhBC within the machinery, it is difficult to determine precisely where the hydrophobic N-terminal signal contacts the machinery.
We speculate that one function of the subunit hydrophobic N-terminal signal might be to trigger opening of the FlhAB-FliPQR export gate, which rests in an energetically favourable closed conformation to maintain the permeability barrier across the bacterial inner membrane (Kuhlen et al., 2018; Butan et al., 2019; Ward et al., 2018; Kuhlen et al., 2020; Bryant and Fraser, 2021). The atomic resolution structure of FliPQR showed that it contains three gating regions (Kuhlen et al., 2018). FliR provides a loop (the R-plug) that sits within the core of the structure. Below this, five copies of FliP each provides three methionine residues that together form a methionine-rich ring (M-gasket) under which ionic interactions between adjacent FliQ subunits hold the base of the structure shut (Q-latch). Mutational and evolutionary analyses have shown that the R-plug, M-gasket, and Q-latch stabilize the closed export gate conformation to maintain the membrane barrier, preventing the leakage of small molecules whilst allowing the passage of substrates into the export channel (Bryant and Fraser, 2021; Hüsing et al., 2021). We have also shown that the FliPQR export gate opens and closes in response to export substrate availability, indicating that the export gate reverts to a closed conformation in the absence of export substrates, thereby maintaining the integrity of the cell membrane (Bryant and Fraser, 2021). These data indicate that the export gate must be opened in response to substrate docking at the export machinery (Bryant and Fraser, 2021; Hüsing et al., 2021).
If the function of the subunit hydrophobic N-terminal signal was to trigger opening of this gate, we hypothesised that mutations which destabilised the gate’s closed conformation would suppress the motility defect associated with FlgDshort, in which the distance between the N-terminal signal and the GRM is reduced. Introduction of the FliP-M210A mutation, which partially destabilises the gate’s closed state, did indeed partly suppress the FlgDshort motility defect. We did not find export gate variants that completely destabilised gate closure, but it may be that such mutations disrupt the membrane permeability barrier (Ward et al., 2018). This could also explain why in screens for suppressors of FlgD∆2–5 or FlgDshort we did not isolate mutations in genes encoding export gate components (data not shown).
The surface-exposed hydrophobic GRM-binding pocket on FlhBC is well conserved across the T3SS SctU family, to which FlhB belongs (Evans et al., 2013; Zarivach et al., 2008; Lountos et al., 2009). Furthermore, the GRM is conserved in all four injectisome early subunits (SctI, SctF, SctP, OrgC) and is located at least 30 residues away from small hydrophobic residues near the subunit N-terminus (Figure 6—figure supplement 4). It therefore seems plausible that the ‘dual signal’ mechanism we propose for early flagellar export operates in all T3SS pathways.
In many other pathways, the presence of a substrate triggers opening or assembly of the export channel. The outer membrane chitin transporter in Vibrio adopts a closed conformation in which the N-terminus of a neighbouring subunit acts as a pore plug (Aunkham et al., 2018). Chitin binding to the transporter ejects the plug, opening the transport channel and allowing chitin transport (Aunkham et al., 2018). In the Sec pathway, interactions of SecA, ribosomes or pre-proteins with SecYEG can induce conformational changes that promote channel opening (Tsirigotaki et al., 2017; Ge et al., 2014; Zimmer et al., 2008; Voorhees et al., 2014). In the TAT system, which transports folded substrates across the cytoplasmic membrane, substrate binding to the TatBC complex triggers association with, and subsequent polymerisation of, TatA, which is required for substrate translocation (Palmer and Berks, 2012; Mori and Cline, 2002). All of these mechanisms serve both to conserve energy and prevent disruption of the membrane permeability barrier. Our data suggest that in a comparable way the signal of non-polar residues within the N-termini of early rod/hook subunits trigger export gate opening.
Materials and methods
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background, (Salmonella enterica serovar Typhimurium) | SJW1103 | doi:10.1099/00221287-130-12-3339 | wildtype | This strain can be obtained from the Fraser lab upon request |
| Strain, strain background, (Salmonella enterica serovar Typhimurium) | recA null | This work | recA gene replaced with kanamycin resistance cassette | This strain can be obtained from the Fraser lab upon request |
| Strain, strain background, (Salmonella enterica serovar Typhimurium) | flgD null | doi:10.1038/nature12682 | flgD gene replaced with kanamycin resistance cassette | This strain can be obtained from the Fraser lab upon request |
| Strain, strain background, (Salmonella enterica serovar Typhimurium) | fliP(M210A)internal HA tag, flgD null | This work | Triple HA tag inserted between residue 21 and 22 of fliP and M210A mutation introduced into the fliP gene, flgD gene replaced with kanamycin resistance cassette. | This strain can be obtained from the Fraser lab upon request |
| Strain, strain background, (Salmonella enterica serovar Typhimurium) | fliPinternal HA tag, flgD null | This work | Triple HA tag inserted between residue 21 and 22 introduced into the fliP gene, flgD gene replaced with kanamycin resistance cassette. | This strain can be obtained from the Fraser lab upon request |
| Recombinant DNA reagent | pTrc99a FlgD | This work | FlgD residues 1-232aa | This vector can be obtained from the Fraser lab upon request |
| Recombinant DNA reagent | pTrc99a FlgD∆2–5 | This work | FlgD residues 1, 6-232aa | This vector can be obtained from the Fraser lab upon request |
| Recombinant DNA reagent | pTrc99a FlgD∆6–10 | This work | FlgD residues 1–5, 11-232aa | This vector can be obtained from the Fraser lab upon request |
| Recombinant DNA reagent | pTrc99a FlgD∆11–15 | This work | FlgD residues 1–10, 16-232aa | This vector can be obtained from the Fraser lab upon request |
| Recombinant DNA reagent | pTrc99a FlgD∆16–20 | This work | FlgD residues 1–15, 21-232aa | This vector can be obtained from the Fraser lab upon request |
| Recombinant DNA reagent | pTrc99a FlgD∆21–25 | This work | FlgD residues 1–20, 26-232aa | This vector can be obtained from the Fraser lab upon request |
| Recombinant DNA reagent | pTrc99a FlgD∆26–30 | This work | FlgD residues 1–25, 31-232aa | This vector can be obtained from the Fraser lab upon request |
| Recombinant DNA reagent | pTrc99a FlgD∆31–35 | This work | FlgD residues 1–30, 36-232aa | This vector can be obtained from the Fraser lab upon request |
| Recombinant DNA reagent | pTrc99a FlgD∆36–40 | This work | FlgD residues 1–35, 41-232aa | This vector can be obtained from the Fraser lab upon request |
| Recombinant DNA reagent | pTrc99a FlgD∆41–45 | This work | FlgD residues 1–40, 46-232aa | This vector can be obtained from the Fraser lab upon request |
| Recombinant DNA reagent | pTrc99a FlgD∆46–50 | This work | FlgD residues 1–45, 51-232aa | This vector can be obtained from the Fraser lab upon request |
| Recombinant DNA reagent | pTrc99a FlgD∆2–5, ∆GRM | This work | FlgD residues 1, 6–35, 41-232aa | This vector can be obtained from the Fraser lab upon request |
| Recombinant DNA reagent | pTrc99a FlgD∆2–5 19(AGAGAG)20 | This work | FlgD residues 1–19, Ala-Gly-Ala-Gly-Ala-Gly, 20-232aa | This vector can be obtained from the Fraser lab upon request |
| Recombinant DNA reagent | pTrc99a FlgD∆2–5 19(STSTST)20 | This work | FlgD residues 1–19, Ser-Thr-Ser-Thr-Ser-Thr, 20-232aa | This vector can be obtained from the Fraser lab upon request |
| Recombinant DNA reagent | pTrc99a FlgD∆2–5 19(GSGSMT)20 | This work | FlgD residues 1–19, Gly-Ser-Gly-Ser-Met-Thr, 20-232aa | This vector can be obtained from the Fraser lab upon request |
| Recombinant DNA reagent | pTrc99a FlgD∆9–32, 4xRpt | This work | FlgD residues 1–8, 4 x(Gly-Ser-Thr-Asn-Ala-Ser), 33-232aa | This vector can be obtained from the Fraser lab upon request |
| Recombinant DNA reagent | pTrc99a FlgD∆9–32, 3xRpt | This work | FlgD residues 1–8, 3 x(Gly-Ser-Thr-Asn-Ala-Ser), 33-232aa | This vector can be obtained from the Fraser lab upon request |
| Recombinant DNA reagent | pTrc99a FlgD∆9–32, 2xRpt (FlgDshort) | This work | FlgD residues 1–8, 2 x(Gly-Ser-Thr-Asn-Ala-Ser), 33-232aa | This vector can be obtained from the Fraser lab upon request |
| Recombinant DNA reagent | pTrc99a FlgD∆9–32, 1xRpt | This work | FlgD residues 1–8, Gly-Ser-Thr-Asn-Ala-Ser, 33-232aa | This vector can be obtained from the Fraser lab upon request |
| Recombinant DNA reagent | pTrc99a FlgD∆9–32 | This work | FlgD residues 1–8, 33-232aa | This vector can be obtained from the Fraser lab upon request |
| Recombinant DNA reagent | pTrc99a FlgD∆9–32, 2xRpt + 1 | This work | FlgD residues 1–8, Gly, 2 x(Gly-Ser-Thr-Asn-Ala-Ser), 33-232aa | This vector can be obtained from the Fraser lab upon request |
| Recombinant DNA reagent | pTrc99a FlgD∆9–32, 2xRpt + 2 | This work | FlgD residues 1–8, Gly-Ser, 2 x(Gly-Ser-Thr-Asn-Ala-Ser), 33-232aa | This vector can be obtained from the Fraser lab upon request |
| Recombinant DNA reagent | pTrc99a FlgD∆9–32, 2xRpt + 3 | This work | FlgD residues 1–8, Gly-Ser-Thr, 2 x(Gly-Ser-Thr-Asn-Ala-Ser), 33-232aa | This vector can be obtained from the Fraser lab upon request |
| Recombinant DNA reagent | pTrc99a FlgD∆9–32, 2xRpt + 4 | This work | FlgD residues 1–8, Gly-Ser-Thr-Asn, 2 x(Gly-Ser-Thr-Asn-Ala-Ser), 33-232aa | This vector can be obtained from the Fraser lab upon request |
| Recombinant DNA reagent | pTrc99a FlgD∆9–32, 2xRpt + 5 | This work | FlgD residues 1–8, Gly-Ser-Thr-Asn-Ala, 2 x(Gly-Ser-Thr-Asn-Ala-Ser), 33-232aa | This vector can be obtained from the Fraser lab upon request |
| Recombinant DNA reagent | pTrc99a FlgDshort, ∆GRM | This work | FlgD residues 1–8, 2 x(Gly-Ser-Thr-Asn-Ala-Ser), 33–35, 41-232aa | This vector can be obtained from the Fraser lab upon request |
| Recombinant DNA reagent | pTrc99a FlgD∆2–5 | This work | FlgD residues 1, 6-232aa | This vector can be obtained from the Fraser lab upon request |
| Recombinant DNA reagent | pTrc99a FlgD∆2–5, M7I | This work | FlgD residues 1, 6-232aa, M7I | This vector can be obtained from the Fraser lab upon request |
| Recombinant DNA reagent | pTrc99a FlgD∆2–5, M7V | This work | FlgD residues 1, 6-232aa, M7V | This vector can be obtained from the Fraser lab upon request |
| Recombinant DNA reagent | pTrc99a FlgD∆2–5, N8I | This work | FlgD residues 1, 6-232aa, N8I | This vector can be obtained from the Fraser lab upon request |
| Recombinant DNA reagent | pTrc99a FlgD∆2–5, D9A | This work | FlgD residues 1, 6-232aa, D9A | This vector can be obtained from the Fraser lab upon request |
| Recombinant DNA reagent | pTrc99a FlgD∆2–5, P10L | This work | FlgD residues 1, 6-232aa, P10L | This vector can be obtained from the Fraser lab upon request |
| Recombinant DNA reagent | pTrc99a FlgD∆2–5, T11I | This work | FlgD residues 1, 6-232aa, T11I | This vector can be obtained from the Fraser lab upon request |
| Recombinant DNA reagent | pTrc99a FlgD∆2–5, 23(TTGSGS)24 | This work | FlgD residues 1–23, Thr-Thr-Gly-Ser-Gly-Ser, 24-232aa | This vector can be obtained from the Fraser lab upon request |
| Recombinant DNA reagent | pTrc99a FlgD∆2–5, 23(TTGSGSTTGSGS)24 | This work | FlgD residues 1–23, Thr-Thr-Gly-Ser-Gly-Ser-Thr-Thr-Gly-Ser-Gly-Ser, 24-232aa | This vector can be obtained from the Fraser lab upon request |
| Recombinant DNA reagent | pTrc99a FlgD∆2–5, 27(GSMTGS)28 | This work | FlgD residues 1–27, Gly-Ser-Met-Thr-Gly-Ser, 28-232aa | This vector can be obtained from the Fraser lab upon request |
| Recombinant DNA reagent | pTrc99a FlgD∆9–32, 8 (2xGSTNAS)33, V15A | This work | FlgD residues 1–8, 2 x(Gly-Ser-Thr-Asn-Ala-Ser), 33-232aa, V15A | This vector can be obtained from the Fraser lab upon request |
| Recombinant DNA reagent | pTrc99a FlgD∆9–32, 8 (2xGSTNAS)33, V15A, M7I | This work | FlgD residues 1–8, 2 x(Gly-Ser-Thr-Asn-Ala-Ser), 33-232aa, M7I | This vector can be obtained from the Fraser lab upon request |
| Recombinant DNA reagent | pTrc99a FlgD∆9–32, 8 (2xGSTNAS)33, V15A, D9A | This work | FlgD residues 1–8, 2 x(Gly-Ser-Thr-Asn-Ala-Ser), 33-232aa, D9A | This vector can be obtained from the Fraser lab upon request |
| Recombinant DNA reagent | pTrc99a FlgD∆9–32, 8 (2xGSTNAS)33, V15A, T11I | This work | FlgD residues 1–8, 2 x(Gly-Ser-Thr-Asn-Ala-Ser), 33-232aa, T11I | This vector can be obtained from the Fraser lab upon request |
| Recombinant DNA reagent | pTrc99a FlgD∆9–32, 8 (2xGSTNAS)33, V15A, G14V | This work | FlgD residues 1–8, 2 x(Gly-Ser-Thr-Asn-Ala-Ser), 33-232aa, G14V | This vector can be obtained from the Fraser lab upon request |
| Recombinant DNA reagent | pTrc99a FlgD∆9–32, 8 (2xGSTNAS-TNPGSTNAS)33 | This work | FlgD residues 1–8, 2 x(Gly-Ser-Thr-Asn-Ala-Ser), (Thr-Asn-Pro-Gly-Ser-Thr-Asn-Ala-Ser) 33-232aa, | This vector can be obtained from the Fraser lab upon request |
| Recombinant DNA reagent | pTrc99a FlgD∆9–32, 8 (2xGSTNAS-GNASGSTNAS)33 | This work | FlgD residues 1–8, 2 x(Gly-Ser-Thr-Asn-Ala-Ser), (Gly-Asn-Ala-Ser-Gly-Ser-Thr-Asn-Ala-Ser) 33-232aa, | This vector can be obtained from the Fraser lab upon request |
| Recombinant DNA reagent | pTrc99a FlgD∆9–32, 8 (2xGSTNAS-QSSFLTLLVAQLKNQDPTNPLQNNELTTQLA)33 | This work | FlgD residues 1–8, 2 x(Gly-Ser-Thr-Asn-Ala-Ser), (Gln-Ser-Ser-Phe-Leu-Thr-Leu-Leu-Val-Ala-Gln-Leu-Lys-Asn-Gln-Asp-Pro-Thr-Asn-Pro-Leu-Asn-Asn-Glu-Leu-Thr-Thr-Gln-Leu-Ala), 33-232aa, | This vector can be obtained from the Fraser lab upon request |
| Recombinant DNA reagent | pTrc99a FlgD∆9–32, 8 (2xGSTNAS-TNASGSTNAS)33 | This work | FlgD residues 1–8, 2 x(Gly-Ser-Thr-Asn-Ala-Ser), (Thr-Asn-Ala-Ser-Gly-Ser-Thr-Asn-Ala-Ser) 33-232aa, | This vector can be obtained from the Fraser lab upon request |
| Recombinant DNA reagent | pTrc99a FlgD∆9–32, 8 (2xGSTNAS-QSSLGSTNAS)34 | This work | FlgD residues 1–8, 2 x(Gly-Ser-Thr-Asn-Ala-Ser), (Gln-Ser-Ser-Leu-Gly-Ser-Thr-Asn-Ala-Ser) 33-232aa, | This vector can be obtained from the Fraser lab upon request |
| Recombinant DNA reagent | pTrc99a FlgD∆9–32, 8 (2xGSTNAS-QNASGSTNAS)35 | This work | FlgD residues 1–8, 2 x(Gly-Ser-Thr-Asn-Ala-Ser), (Gln-Asn-Ala-Ser-Gly-Ser-Thr-Asn-Ala-Ser) 33-232aa, | This vector can be obtained from the Fraser lab upon request |
| Recombinant DNA reagent | pTrc99a FlgD∆9–32, 8 (2xGSTNAS-TNTFGTLIAS)36 | This work | FlgD residues 1–8, 2 x(Gly-Ser-Thr-Asn-Ala-Ser), (Thr-Asn-Thr-Phe-Gly-Thr-Leu-Iso-Ala-Ser) 33-232aa, | This vector can be obtained from the Fraser lab upon request |
| Recombinant DNA reagent | pTrc99a FlgG | This work | FlgG residues 1–144, FLAGx3, 145-260aa | This vector can be obtained from the Fraser lab upon request |
| Recombinant DNA reagent | pTrc99a FlgG∆short | This work | FlgG residues 1–10, 35–144, FLAGx3, 145-260aa | This vector can be obtained from the Fraser lab upon request |
| Recombinant DNA reagent | pTrc99a FlgGshort+ linker | This work | FlgG residues 1–10, 4 x(Gly-Ser-Thr-Asn-Ala-Ser) 35–144, FLAGx3, 145-260aa | This vector can be obtained from the Fraser lab upon request |
| Recombinant DNA reagent | pTrc99a FlgE | This work | FlgE residues 1–234, FLAGx3, 235-403aa | This vector can be obtained from the Fraser lab upon request |
| Recombinant DNA reagent | pTrc99a FlgEshort | This work | FlgE residues 1–8, 33–234, FLAGx3, 235-403aa | This vector can be obtained from the Fraser lab upon request |
| Recombinant DNA reagent | pTrc99a FlgEshort+ linker | This work | FlgE residues 1–8, 4 x(Gly-Ser-Thr-Asn-Ala-Ser) 33–234, FLAGx3, 235-403aa | This vector can be obtained from the Fraser lab upon request |
| Recombinant DNA reagent | pTrc99a FlgE∆GRM | This work | FlgE residues 1–38, 44–234, FLAGx3, 235-403aa | This vector can be obtained from the Fraser lab upon request |
| Recombinant DNA reagent | pTrc99a FlgEshort, ∆GRM | This work | FlgE residues 1–8, 33–38, 44–234, FLAGx3, 235-403aa | This vector can be obtained from the Fraser lab upon request |
| Recombinant DNA reagent | pTrc99a FliKmyc | This work | FliK residues 1-405aa, myc | This vector can be obtained from the Fraser lab upon request |
| Recombinant DNA reagent | pTrc99a FliKmyc∆2–8 | This work | FliK residues 1, 9-405aa, myc | This vector can be obtained from the Fraser lab upon request |
| Antibody | anti-FLAG (Mouse monoclonal) | Sigma-Aldrich | Cat# F3165, RRID:AB_259529 | Mouse monoclonal against FLAG tag (1:1000) |
| Antibody | Anti-HA Tag, HRP conjugate(Mouse monoclonal) | Thermo Fisher Scientific | Cat # 26183-HRP, RRID:AB_2533056 | Mouse monoclonal against HA tag (1:1000) |
| Antibody | anti-Myc (9B11), HRP conjugate (Mouse monoclonal) | Cell signalling technology | Cat # 2040, RRID:AB_2148465 | Mouse monoclonal against Myc tag (1:1000) |
| Antibody | anti-FlgD (Rabbit polyclonal) | doi:10.1038/nature12682 | Rabbit polyclonal against Salmonella FlgD (1:1000). This antibody can be obtained from the Fraser lab upon request. | |
| Antibody | anti-FliK (Rabbit polyclonal) | This work | Rabbit polyclonal against Salmonella FliK (1:1000). This antibody can be obtained from the Fraser lab upon request. | |
| Antibody | anti-FlgK (Rabbit polyclonal) | doi: 10.1111/mmi.14731 | Rabbit polyclonal against Salmonella FlgK (1:1000). This antibody can be obtained from the Fraser lab upon request. | |
| Antibody | anti-FlgL (Rabbit polyclonal) | This work | Rabbit polyclonal against Salmonella FlgL (1:1000). This antibody can be obtained from the Fraser lab upon request. | |
| Antibody | anti-FlgN (Rabbit polyclonal) | doi: 10.1111/mmi.14731 | Rabbit polyclonal against Salmonella FlgN (1:1000). This antibody can be obtained from the Fraser lab upon request. | |
| Antibody | anti-FlhA (Rabbit polyclonal) | doi: 10.1111/mmi.14731 | Rabbit polyclonal against Salmonella FlhA (1:1000). This antibody can be obtained from the Fraser lab upon request. |
Bacterial strains, plasmids, and growth conditions
Request a detailed protocolSalmonella strains and plasmids used in this study are listed in Table 1. The ΔflgD::KmR strain in which the flgD gene was replaced by a kanamycin resistance cassette was constructed using the λ Red recombinase system (Datsenko and Wanner, 2000). Strains containing chromosomally encoded FliP variants were constructed by aph-I-SceI Kanamycin resistance cassette replacement using pWRG730 (Hoffmann et al., 2017). Recombinant proteins were expressed in Salmonella from the isopropyl β-D-thiogalactoside-inducible (IPTG) inducible plasmid pTrc99a (Amann et al., 1988). Bacteria were cultured at 30–37°C in Luria-Bertani (LB) broth containing ampicillin (100 μg/ml).
Strains and recombinant plasmids.
| Strains | Description |
|---|---|
| Salmonella typhimurium | |
| SJW1103 | wildtype |
| recA null | ∆recA::kmR |
| flgD null | ∆flgD::kmR |
| fliP(M210A)internalHAtag, flgD null | fliP(M210A) 21 (3xHA tag)22, ∆flgD::kmR |
| fliPinternalHAtag, flgD null | fliP 21 (3xHA tag)22, ∆flgD::kmR |
| Plasmids | |
| pTrc99a FlgD | 1-232aa |
| pTrc99a FlgD∆2–5 | 1, 6-232aa |
| pTrc99a FlgD∆6–10 | 1–5, 11-232aa |
| pTrc99a FlgD∆11–15 | 1–10, 16-232aa |
| pTrc99a FlgD∆16–20 | 1–15, 21-232aa |
| pTrc99a FlgD∆21–25 | 1–20, 26-232aa |
| pTrc99a FlgD∆26–30 | 1–25, 31-232aa |
| pTrc99a FlgD∆31–35 | 1–30, 36-232aa |
| pTrc99a FlgD∆36–40 | 1–35, 41-232aa |
| pTrc99a FlgD∆41–45 | 1–40, 46-232aa |
| pTrc99a FlgD∆46–50 | 1–45, 51-232aa |
| pTrc99a FlgD∆2–5, ∆GRM | 1, 6–35, 41-232aa |
| pTrc99a FlgD∆2–5 19(AGAGAG)20 | 1–19, Ala-Gly-Ala-Gly-Ala-Gly, 20-232aa |
| pTrc99a FlgD∆2–5 19(STSTST)20 | 1–19, Ser-Thr-Ser-Thr-Ser-Thr, 20-232aa |
| pTrc99a FlgD∆2–5 19(GSGSMT)20 | 1–19, Gly-Ser-Gly-Ser-Met-Thr, 20-232aa |
| pTrc99a FlgD∆9–32, 4xRpt | 1–8, 4 x(Gly-Ser-Thr-Asn-Ala-Ser), 33-232aa |
| pTrc99a FlgD∆9–32, 3xRpt | 1–8, 3 x(Gly-Ser-Thr-Asn-Ala-Ser), 33-232aa |
| pTrc99a FlgD∆9–32, 2xRpt (FlgDshort) | 1–8, 2 x(Gly-Ser-Thr-Asn-Ala-Ser), 33-232aa |
| pTrc99a FlgD∆9–32, 1xRpt | 1–8, Gly-Ser-Thr-Asn-Ala-Ser, 33-232aa |
| pTrc99a FlgD∆9–32 | 1–8, 33-232aa |
| pTrc99a FlgD∆9–32, 2xRpt + 1 | 1–8, Gly, 2 x(Gly-Ser-Thr-Asn-Ala-Ser), 33-232aa |
| pTrc99a FlgD∆9–32, 2xRpt + 2 | 1–8, Gly-Ser, 2 x(Gly-Ser-Thr-Asn-Ala-Ser), 33-232aa |
| pTrc99a FlgD∆9–32, 2xRpt + 3 | 1–8, Gly-Ser-Thr, 2 x(Gly-Ser-Thr-Asn-Ala-Ser), 33-232aa |
| pTrc99a FlgD∆9–32, 2xRpt + 4 | 1–8, Gly-Ser-Thr-Asn, 2 x(Gly-Ser-Thr-Asn-Ala-Ser), 33-232aa |
| pTrc99a FlgD∆9–32, 2xRpt + 5 | 1–8, Gly-Ser-Thr-Asn-Ala, 2 x(Gly-Ser-Thr-Asn-Ala-Ser), 33-232aa |
| pTrc99a FlgDshort, ∆GRM | 1–8, 2 x(Gly-Ser-Thr-Asn-Ala-Ser), 33–35, 41-232aa |
| pTrc99a FlgD∆2–5 | 1, 6-232aa |
| pTrc99a FlgD∆2–5, M7I | 1, 6-232aa, M7I |
| pTrc99a FlgD∆2–5, M7V | 1, 6-232aa, M7V |
| pTrc99a FlgD∆2–5, N8I | 1, 6-232aa, N8I |
| pTrc99a FlgD∆2–5, D9A | 1, 6-232aa, D9A |
| pTrc99a FlgD∆2–5, P10L | 1, 6-232aa, P10L |
| pTrc99a FlgD∆2–5, T11I | 1, 6-232aa, T11I |
| pTrc99a FlgD∆2–5, 23(TTGSGS)24 | 1–23, Thr-Thr-Gly-Ser-Gly-Ser, 24-232aa |
| pTrc99a FlgD∆2–5, 23(TTGSGSTTGSGS)24 | 1–23, Thr-Thr-Gly-Ser-Gly-Ser-Thr-Thr-Gly-Ser-Gly-Ser, 24-232aa |
| pTrc99a FlgD∆2–5, 27(GSMTGS)28 | 1–27, Gly-Ser-Met-Thr-Gly-Ser, 28-232aa |
| pTrc99a FlgD∆9–32, 8(2xGSTNAS)33, V15A | 1–8, 2 x(Gly-Ser-Thr-Asn-Ala-Ser), 33-232aa, V15A |
| pTrc99a FlgD∆9–32, 8(2xGSTNAS)33, V15A, M7I | 1–8, 2 x(Gly-Ser-Thr-Asn-Ala-Ser), 33-232aa, M7I |
| pTrc99a FlgD∆9–32, 8(2xGSTNAS)33, V15A, D9A | 1–8, 2 x(Gly-Ser-Thr-Asn-Ala-Ser), 33-232aa, D9A |
| pTrc99a FlgD∆9–32, 8(2xGSTNAS)33, V15A, T11I | 1–8, 2 x(Gly-Ser-Thr-Asn-Ala-Ser), 33-232aa, T11I |
| pTrc99a FlgD∆9–32, 8(2xGSTNAS)33, V15A, G14V | 1–8, 2 x(Gly-Ser-Thr-Asn-Ala-Ser), 33-232aa, G14V |
| pTrc99a FlgD∆9–32, 8(2xGSTNAS-TNPGSTNAS)33 | 1–8, 2 x(Gly-Ser-Thr-Asn-Ala-Ser), (Thr-Asn-Pro-Gly-Ser-Thr-Asn-Ala-Ser) 33-232aa, |
| pTrc99a FlgD∆9–32, 8(2xGSTNAS-GNASGSTNAS)33 | 1–8, 2 x(Gly-Ser-Thr-Asn-Ala-Ser), (Gly-Asn-Ala-Ser-Gly-Ser-Thr-Asn-Ala-Ser) 33-232aa, |
| pTrc99a FlgD∆9–32, 8(2xGSTNAS-QSSFLTLLVAQLKNQDPTNPLQNNELTTQLA)33 | 1–8, 2 x(Gly-Ser-Thr-Asn-Ala-Ser), (Gln-Ser-Ser-Phe-Leu-Thr-Leu-Leu-Val-Ala-Gln-Leu-Lys-Asn-Gln-Asp-Pro-Thr-Asn-Pro-Leu-Asn-Asn-Glu-Leu-Thr-Thr-Gln-Leu-Ala), 33-232aa, |
| pTrc99a FlgD∆9–32, 8(2xGSTNAS-TNASGSTNAS)33 | 1–8, 2 x(Gly-Ser-Thr-Asn-Ala-Ser), (Thr-Asn-Ala-Ser-Gly-Ser-Thr-Asn-Ala-Ser) 33-232aa, |
| pTrc99a FlgD∆9–32, 8(2xGSTNAS-QSSLGSTNAS)34 | 1–8, 2 x(Gly-Ser-Thr-Asn-Ala-Ser), (Gln-Ser-Ser-Leu-Gly-Ser-Thr-Asn-Ala-Ser) 33-232aa, |
| pTrc99a FlgD∆9–32, 8(2xGSTNAS-QNASGSTNAS)35 | 1–8, 2 x(Gly-Ser-Thr-Asn-Ala-Ser), (Gln-Asn-Ala-Ser-Gly-Ser-Thr-Asn-Ala-Ser) 33-232aa, |
| pTrc99a FlgD∆9–32, 8(2xGSTNAS-TNTFGTLIAS)36 | 1–8, 2 x(Gly-Ser-Thr-Asn-Ala-Ser), (Thr-Asn-Thr-Phe-Gly-Thr-Leu-Iso-Ala-Ser) 33-232aa, |
| pTrc99a FlgG | 1–144, FLAGx3, 145-260aa |
| pTrc99a FlgG∆short | 1–10, 35–144, FLAGx3, 145-260aa |
| pTrc99a FlgGshort+ linker | 1–10, 4 x(Gly-Ser-Thr-Asn-Ala-Ser) 35–144, FLAGx3, 145-260aa |
| pTrc99a FlgE | 1–234, FLAGx3, 235-403aa |
| pTrc99a FlgEshort | 1–8, 33–234, FLAGx3, 235-403aa |
| pTrc99a FlgEshort+ linker | 1–8, 4 x(Gly-Ser-Thr-Asn-Ala-Ser) 33–234, FLAGx3, 235-403aa |
| pTrc99a FlgE∆GRM | 1–38, 44–234, FLAGx3, 235-403aa |
| pTrc99a FlgEshort, ∆GRM | 1–8, 33–38, 44–234, FLAGx3, 235-403aa |
| pTrc99a FliKmyc | 1-405aa, myc |
| pTrc99a FliKmyc∆2–8 | 1, 9-405aa, myc |
Flagellar subunit export assay
Request a detailed protocolSalmonella strains were cultured at 37 °C in LB broth containing ampicillin and IPTG to mid-log phase (OD600nm 0.6–0.8). Cells were centrifuged (6000 x g, 3 min) and resuspended in fresh media and grown for a further 60 min at 37 °C. The cells were pelleted by centrifugation (16,000 x g, 5 min) and the supernatant passed through a 0.2 μm nitrocellulose filter. Proteins were precipitated with 10% trichloroacetic acid (TCA) and 1% Triton X-100 on ice for 1 hr, pelleted by centrifugation (16,000 x g, 10 min), washed with ice-cold acetone and resuspended in SDS-PAGE loading buffer (volumes calibrated according to cell densities). Fractions were analysed by immunoblotting.
Motility assays
Request a detailed protocolFor swimming motility, cultures were grown in LB broth to A600nm 1. Two microliters of culture were inoculated into soft tryptone agar (0.3% agar, 10 g/L tryptone, 5 g/L NaCl) containing ampicillin (100 μg/ml). Plates were incubated at 37 °C for between 4 and 6 hr unless otherwise stated.
Isolation of motile strains carrying suppressor mutations
Request a detailed protocolCells of the Salmonella flgD null strain transformed with plasmids expressing FlgD variants (FlgDΔ2–5, FlgDΔ2-5-19GSGSMT20-V15A or FlgDshort) were cultured at 37 °C in LB broth containing ampicillin (100 μg/ml) to mid-log phase and inoculated into soft tryptone agar (0.3% agar, 10 g/L tryptone, 5 g/L NaCl) containing ampicillin (100 μg/ml). Plates were incubated at 30 °C until motile ‘spurs’ appeared. Cells from the spurs were streaked to single colony and cultured to isolate the flgD encoding plasmid. Plasmids were transformed into the Salmonella flgD null strain to assess whether the plasmids were responsible for the motile suppressor phenotypes. Plasmids were sequenced to identify the suppressor mutations.
Quantification and statistical analysis
Request a detailed protocolExperiments were performed at least three times. Immunoblots were quantified using Image Studio Lite. The unpaired two-tailed Student’s t-test was used to determine p-values and significance was determined as *p< 0.05. Data are represented as mean ± standard error of the mean (SEM), unless otherwise specified and reported as biological replicates.
Data availability
Data generated or analysed during this study are included in the manuscript and supporting files or have been submitted to Dryad (https://doi.org/10.5061/dryad.66t1g1k3x).
-
Dryad Digital RepositoryData from: Recognition of discrete export signals in early flagellar subunits during bacterial Type III secretion.https://doi.org/10.5061/dryad.66t1g1k3x
References
-
Architecture of the major component of the type III secretion system export apparatusNature Structural & Molecular Biology 20:99–104.https://doi.org/10.1038/nsmb.2452
-
Termini of Salmonella flagellin are disordered and become organized upon polymerization into flagellar filamentJournal of Molecular Biology 211:673–677.https://doi.org/10.1016/0022-2836(90)90064-S
-
Structural basis for chitin acquisition by marine Vibrio speciesNature Communications 9:220.https://doi.org/10.1038/s41467-017-02523-y
-
Chaperone-mediated coupling of subunit availability to activation of flagellar Type III secretionMolecular Microbiology 116:538–549.https://doi.org/10.1111/mmi.14731
-
Type III protein secretion in plant pathogenic bacteriaPlant Physiology 150:1656–1664.https://doi.org/10.1104/pp.109.139089
-
Corrigendum: Assembly, structure, function and regulation of type III secretion systemsNature Reviews. Microbiology 15:379.https://doi.org/10.1038/nrmicro.2017.54
-
Building a flagellum in biological outer spaceMicrobial Cell (Graz, Austria) 1:64–66.https://doi.org/10.15698/mic2014.01.128
-
Building a flagellum outside the bacterial cellTrends in Microbiology 22:566–572.https://doi.org/10.1016/j.tim.2014.05.009
-
Control of membrane barrier during bacterial type-III protein secretionNature Communications 12:3999.https://doi.org/10.1038/s41467-021-24226-1
-
Interaction between FliJ and FlhA, Components of the Bacterial Flagellar Type III Export ApparatusJournal of Bacteriology 195:466–473.https://doi.org/10.1128/JB.01711-12
-
Structure of the core of the type III secretion system export apparatusNature Structural & Molecular Biology 25:583–590.https://doi.org/10.1038/s41594-018-0086-9
-
A simple method for displaying the hydropathic character of A proteinJournal of Molecular Biology 157:105–132.https://doi.org/10.1016/0022-2836(82)90515-0
-
Inhibition of a type III secretion system by the deletion of a short loop in one of its membrane proteinsActa Crystallographica. Section D, Biological Crystallography 69:812–820.https://doi.org/10.1107/S0907444913002102
-
Components of the Salmonella Flagellar Export Apparatus and Classification of Export SubstratesJournal of Bacteriology 181:1388–1394.https://doi.org/10.1128/JB.181.5.1388-1394.1999
-
The twin-arginine translocation (Tat) protein export pathwayNature Reviews. Microbiology 10:483–496.https://doi.org/10.1038/nrmicro2814
-
Protein export through the bacterial Sec pathwayNature Reviews. Microbiology 15:21–36.https://doi.org/10.1038/nrmicro.2016.161
-
Localization of the flagellum-specific secretion signal in Salmonella flagellinBiochemical and Biophysical Research Communications 345:93–98.https://doi.org/10.1016/j.bbrc.2006.04.055
-
Terminal disorder: A common structural feature of the axial proteins of bacterial flagellum?Journal of Molecular Biology 226:575–579.https://doi.org/10.1016/0022-2836(92)90616-r
-
Type-III secretion pore formed by flagellar protein FliPMolecular Microbiology 107:94–103.https://doi.org/10.1111/mmi.13870
-
Mutations in fliK and flhB affecting flagellar hook and filament assembly in Salmonella typhimuriumJournal of Bacteriology 178:2960–2970.https://doi.org/10.1128/jb.178.10.2960-2970.1996
Article and author information
Author details
Funding
Biotechnology and Biological Sciences Research Council (BB/M007197/1)
- Gillian M Fraser
The Wellcome Trust
- Colin Hughes
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Acknowledgements
This work was funded by grants from the Biotechnology and Biological Sciences Research Council (BB/M007197/1) to GMF, the Wellcome Trust (082895/Z/07/Z) to CH and GMF, a Biotechnology and Biological Sciences Research Council studentship to P.D., and a University of Cambridge John Lucas Walker studentship to OJB.
Version history
- Preprint posted: December 9, 2020 (view preprint)
- Received: January 5, 2021
- Accepted: March 2, 2022
- Accepted Manuscript published: March 3, 2022 (version 1)
- Version of Record published: April 5, 2022 (version 2)
Copyright
© 2022, Bryant et al.
This article is distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use and redistribution provided that the original author and source are credited.
Metrics
-
- 939
- views
-
- 173
- downloads
-
- 7
- citations
Views, downloads and citations are aggregated across all versions of this paper published by eLife.
Download links
Downloads (link to download the article as PDF)
Open citations (links to open the citations from this article in various online reference manager services)
Cite this article (links to download the citations from this article in formats compatible with various reference manager tools)
Further reading
-
- Microbiology and Infectious Disease
The agr quorum-sensing system links Staphylococcus aureus metabolism to virulence, in part by increasing bacterial survival during exposure to lethal concentrations of H2O2, a crucial host defense against S. aureus. We now report that protection by agr surprisingly extends beyond post-exponential growth to the exit from stationary phase when the agr system is no longer turned on. Thus, agr can be considered a constitutive protective factor. Deletion of agr resulted in decreased ATP levels and growth, despite increased rates of respiration or fermentation at appropriate oxygen tensions, suggesting that Δagr cells undergo a shift towards a hyperactive metabolic state in response to diminished metabolic efficiency. As expected from increased respiratory gene expression, reactive oxygen species (ROS) accumulated more in the agr mutant than in wild-type cells, thereby explaining elevated susceptibility of Δagr strains to lethal H2O2 doses. Increased survival of wild-type agr cells during H2O2 exposure required sodA, which detoxifies superoxide. Additionally, pretreatment of S. aureus with respiration-reducing menadione protected Δagr cells from killing by H2O2. Thus, genetic deletion and pharmacologic experiments indicate that agr helps control endogenous ROS, thereby providing resilience against exogenous ROS. The long-lived ‘memory’ of agr-mediated protection, which is uncoupled from agr activation kinetics, increased hematogenous dissemination to certain tissues during sepsis in ROS-producing, wild-type mice but not ROS-deficient (Cybb−/−) mice. These results demonstrate the importance of protection that anticipates impending ROS-mediated immune attack. The ubiquity of quorum sensing suggests that it protects many bacterial species from oxidative damage.
-
- Medicine
- Microbiology and Infectious Disease
Background:
End-stage renal disease (ESRD) patients experience immune compromise characterized by complex alterations of both innate and adaptive immunity, and results in higher susceptibility to infection and lower response to vaccination. This immune compromise, coupled with greater risk of exposure to infectious disease at hemodialysis (HD) centers, underscores the need for examination of the immune response to the COVID-19 mRNA-based vaccines.
Methods:
The immune response to the COVID-19 BNT162b2 mRNA vaccine was assessed in 20 HD patients and cohort-matched controls. RNA sequencing of peripheral blood mononuclear cells was performed longitudinally before and after each vaccination dose for a total of six time points per subject. Anti-spike antibody levels were quantified prior to the first vaccination dose (V1D0) and 7 d after the second dose (V2D7) using anti-spike IgG titers and antibody neutralization assays. Anti-spike IgG titers were additionally quantified 6 mo after initial vaccination. Clinical history and lab values in HD patients were obtained to identify predictors of vaccination response.
Results:
Transcriptomic analyses demonstrated differing time courses of immune responses, with prolonged myeloid cell activity in HD at 1 wk after the first vaccination dose. HD also demonstrated decreased metabolic activity and decreased antigen presentation compared to controls after the second vaccination dose. Anti-spike IgG titers and neutralizing function were substantially elevated in both controls and HD at V2D7, with a small but significant reduction in titers in HD groups (p<0.05). Anti-spike IgG remained elevated above baseline at 6 mo in both subject groups. Anti-spike IgG titers at V2D7 were highly predictive of 6-month titer levels. Transcriptomic biomarkers after the second vaccination dose and clinical biomarkers including ferritin levels were found to be predictive of antibody development.
Conclusions:
Overall, we demonstrate differing time courses of immune responses to the BTN162b2 mRNA COVID-19 vaccination in maintenance HD subjects comparable to healthy controls and identify transcriptomic and clinical predictors of anti-spike IgG titers in HD. Analyzing vaccination as an in vivo perturbation, our results warrant further characterization of the immune dysregulation of ESRD.
Funding:
F30HD102093, F30HL151182, T32HL144909, R01HL138628. This research has been funded by the University of Illinois at Chicago Center for Clinical and Translational Science (CCTS) award UL1TR002003.






