USP13 antagonizes gp78 to maintain functionality of a chaperone in ER-associated degradation
Figures
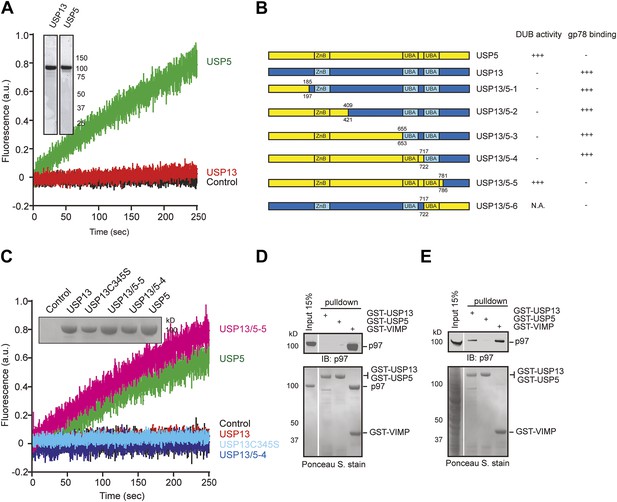
USP13 and USP5 have distinct activities despite sequence homology.
(A) Purified USP13 is inactive whereas USP5 is active. The activities of the purified USP5 and USP13 were measured by the Ub-AFC assay. The Coomassie blue-stained gels show the purified proteins. (B) A schematic illustration of the USP13-USP5 chimeras and a summary of their deubiquitinating activities. (C) The deubiquitinating activity of the indicated USP13-USP5 chimeras as measured by the Ub-AFC assay. The USP13 catalytic inactive mutant C345S was used as a negative control. The inset shows the purified proteins. (D) USP13 does not bind p97 directly. The indicated GST-tagged proteins were immobilized and incubated with recombinant p97. The precipitated proteins were analyzed by immunoblotting (IB). (E) USP13, but not USP5, binds p97 through an adaptor. As in D, except that a whole cell extract was used in replace of p97.
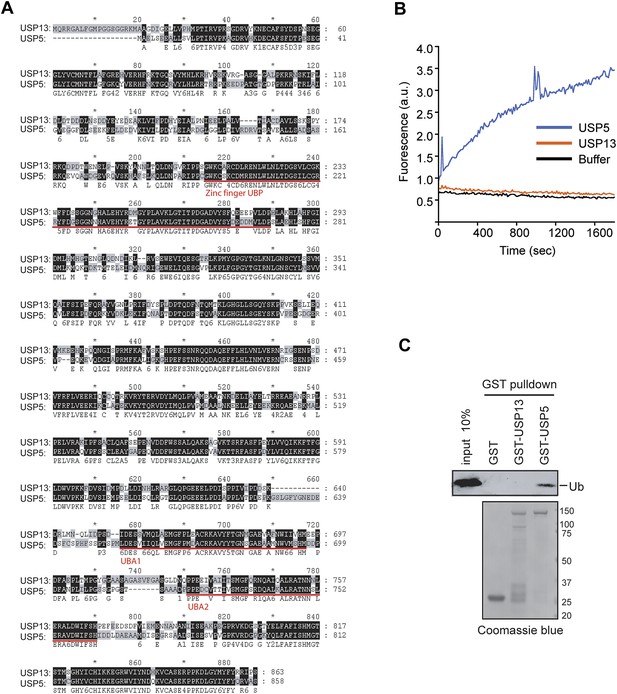
USP13 and USP5 have distinct deubiquitinating activities.
(A) Sequence alignment of USP13 and USP5. (B) Deubiquitinating activity of recombinant USP13 and USP5 purified from E. coli. Purified USP5 and USP13 (200 nM) were incubated with Ub-AFC (0.5 µM) and the fluorescence intensity was measured. (C) USP13 does not bind free ubiquitin directly. The indicated GST-tagged proteins were immobilized and incubated with ubiquitin. The precipitated proteins were analyzed by SDS-PAGE and Coomassie blue staining.
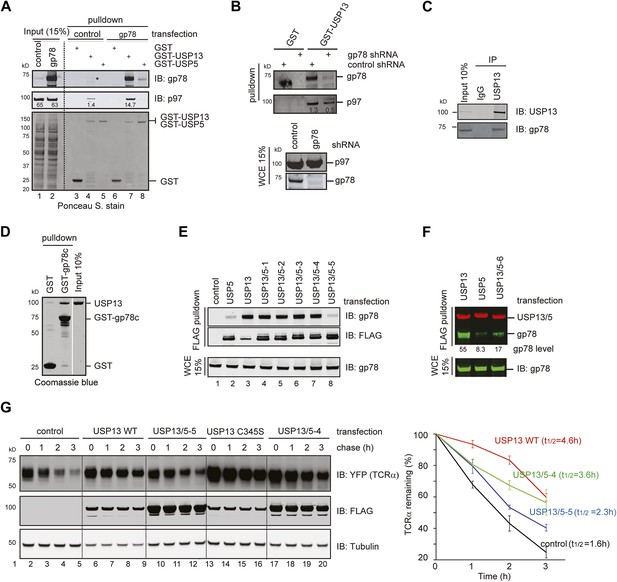
USP13 forms a complex with the ERAD E3 gp78.
(A) gp78 enhances the interaction of USP13 with p97. Cell extracts prepared from control or gp78-expressing cells were incubated with glutathione beads containing the indicated proteins. The asterisk indicates a fraction of endogenous gp78 that is co-precipitated with GST-USP13. The numbers indicate the levels of p97. (B) Depletion of gp78 reduces the interaction of USP13 with p97. Whole cell extracts (WCE) from control or gp78 knockdown cells were incubated with glutathione beads containing either GST or GST-USP13. (C) An interaction between endogenous USP13 and gp78. Whole cell extract was subject to immunoprecipitation (IP) with either control or anti-USP13 antibodies. (D) USP13 binds the gp78 cytosolic domain (gp78c) directly. The indicated GST-tagged proteins were immobilized and incubated with recombinant USP13. (E and F) Interactions of the indicated USP13-USP5 chimeras with gp78. WCEs from cells expressing the indicated DUB chimeras and gp78 were subject to IP with FLAG beads. (G) ERAD inhibition by overexpressed USP13 is dependent on gp78 interaction. TCRα-YFP cells transfected with the indicated USP13-expressing plasmids were treated with cycloheximide for the indicated time points. Cell extracts were analyzed by immunoblotting. The graph represents quantification of three independent experiments. Error bars, SD (n = 3).
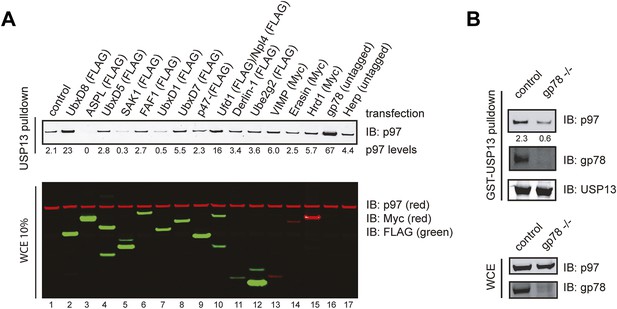
Identification of factors that influence the USP13-p97 interaction.
(A) Cells expressing the indicated proteins were lysed. Cell extracts were incubated with glutathione beads containing GST-USP13. The precipitated materials and a fraction of the input were analyzed by immunoblotting with the indicated antibodies. (B) Depletion of gp78 reduces the interaction of USP13 with p97. Whole cell extracts (WCE) from control or gp78 knockout cells (−/−) were incubated with glutathione beads containing either GST or GST-USP13.
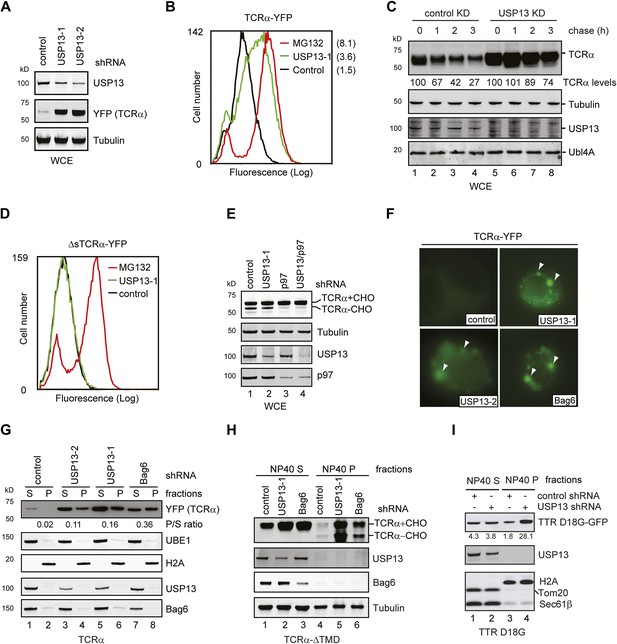
USP13 is required to maintain the solubility of a retrotranslocation substrate.
(A–C) USP13 knockdown stabilizes the ERAD substrate TCRα. (A) HEK293 cells stably expressing TCRα-YFP were transfected with the indicated shRNA-expressing plasmids. Proteins in the whole cell extracts (WCE) were analyzed by immunoblotting. (B) Control or USP13 knockdown cells expressing TCRα-YFP were analyzed by flow cytometry. The proteasome inhibitor MG132 was used as a positive control. The numbers indicate the calculated X-mean values. (C) The stability of TCRα-YFP in control and USP13 knockdown cells was analyzed by CHX chase. The number indicated the relative amount of TCRα-YFP averaged from two independent experiments. (D) USP13 knockdown does not stabilize a non-ERAD substrate. As in B, except that cells stably expressing ΔsTCRα-YFP was used. (E) USP13 functions at a step downstream of p97-mediated dislocation. The TCRα-YFP-expressing cells treated with the indicated shRNA-expressing plasmids were exposed to the proteasome inhibitor MG132 (10 µM, 15 hr). TCRα+CHO, glycosylated TCRα; TCRα-CHO, deglycosylated TCRα. (F) TCRα-YFP-containing aggregates accumulate in USP13- and Bag6-depleted HEK293 cells. Shown are representative cells transfected with a TCRα-YFP-expressing plasmid together with the indicated shRNA constructs. Arrowheads indicate TCRα-YFP punctae formed upon depletion of USP13 or Bag6. (G) TCRα-YFP-containing aggregates in USP13-depleted cells are resistant to extraction by NP40. As in F, except that cells were subject to sequential extraction, first by an NP40-containing lysis buffer, then by the Laemmli buffer. UBE1 and H2A serve as the markers for the NP40 soluble (S) and insoluble (P) fractions, respectively. Note that the NP40-insoluble fractions were loaded two times of the soluble fractions. The numbers indicate the ratio of TCRα in the pellet vs the soluble fraction. (H and I) As in G, except that cells expressing the indicated ERAD substrates were used. The numbers in I indicate the levels of TTR D18G. Note that a Bag6 antibody against the C-terminus of Bag6 was used in G and H.
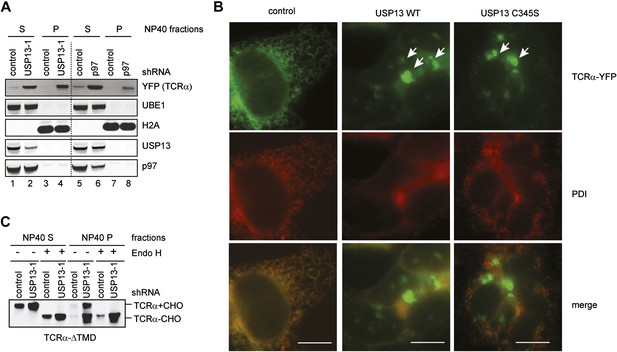
USP13 loss-of-function causes TCRα to accumulate in aggregates in cells.
(A) Cells co-expressing TCRα-YFP together with the indicated shRNAs were subject to sequential extraction, first by an NP40-containing lysis buffer, then by the Laemmli buffer. The NP40 soluble (S) and insoluble (P) fractions were analyzed by immunoblotting. UBE1 and H2A serve as markers for the NP40 soluble (S) and insoluble (P) fractions, respectively. (B) Expression of USP13 inhibits TCRα degradation in a dominant negative manner. COS7 cells transfected with TCRα-YFP together with either control or the indicated USP13-expressing constructs were stained by a PDI antibody in red. Cells were imaged using a Zeiss Axiovert fluorescence microscope with the exposure time set to auto. Note that in USP13 and USP13 C345S transfected cells, TCRα-YFP often accumulates in aggregates that are not localized to the same focal plane as the ER marker PDI. Scale bars, 10 µm. (C) USP13 knockdown stabilizes an ERAD substrate in both glycosylated and deglycosylated forms. Cells co-expressing TCRα-ΔTMD together with the indicated shRNAs were subject to sequential extraction, first by an NP40-containing lysis buffer, then by the Laemmli buffer. Where indicated, the extracts were treated with EndoH before immunoblotting.
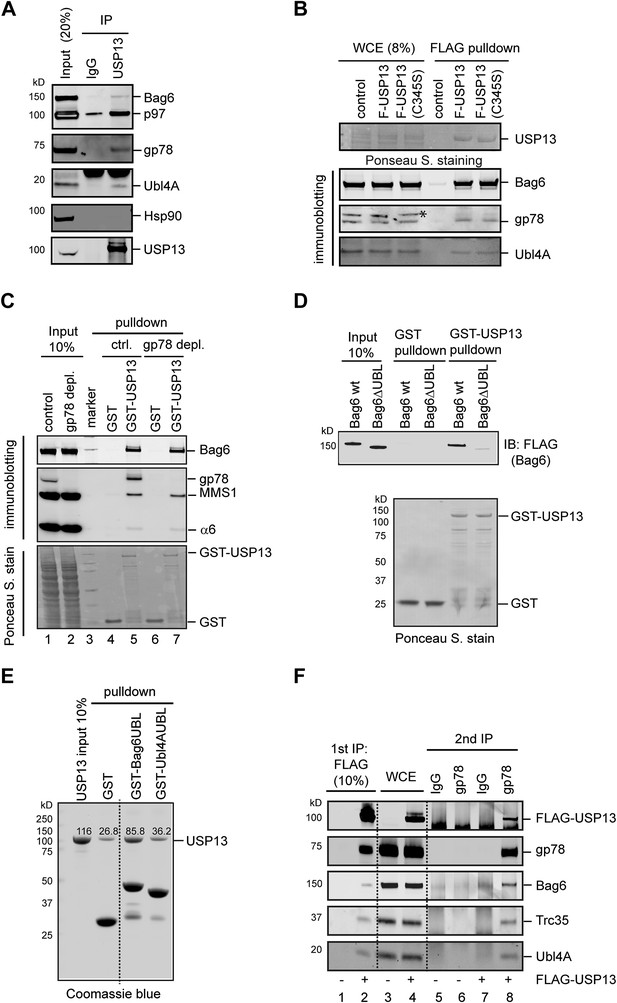
USP13 interacts with the Bag6 complex.
(A) Endogenous interaction of USP13 with the Bag6 complex. Extracts from cells were subject to IP by the indicated antibodies followed by immunoblotting. (B) USP13 binds gp78 and Bag6 independent of its DUB activity. Extracts from HEK293 cells-transfected with the indicated plasmids were subject to IP with anti-FLAG beads. Asterisk indicates a non-specific band. (C) USP13 interacts with both the proteasome and Bag6 in a gp78 independent manner. Control (ctrl.) or gp78-depleted cell extracts were incubated with glutathione beads containing either GST or GST-USP13. The precipitated materials were analyzed by immunoblotting (Upper two panels) or by Ponceau S staining (the lower panel). (D) The UBL domain in Bag6 is required for interaction with USP13. A GST pull-down experiment was performed using the indicated Bag6 proteins purified from mammalian cells. (E) Bag6 UBL binds USP13 more tightly than Ubl4A UBL. Recombinant proteins purified from E. coli were used. (F) USP13, gp78 and Bag6 form a multi-protein complex. Sequential IP first by FLAG beads then by gp78 antibody using extracts from either control (−) or FLAG-USP13 (F-USP13)-expressing cells (indicated by ‘+’).
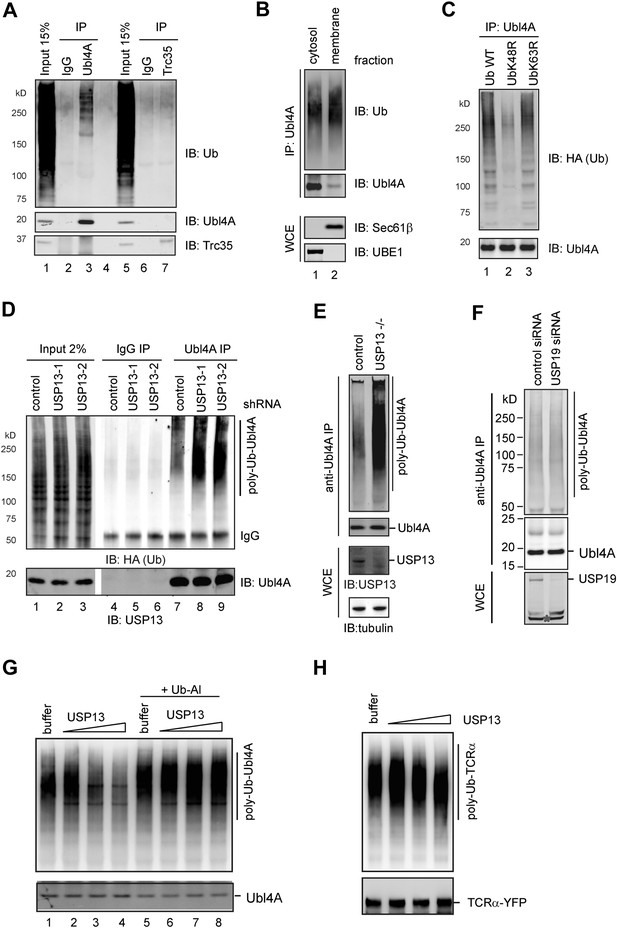
USP13 regulates ubiquitination of Ubl4A A, Ubl4A but not Trc35 is ubiquitinated in cells.
(A) Ubl4A or Trc35 was immunoprecipitated from an extract of HEK293 cells under denaturing condition and blotted with the indicated antibodies. (B) Membrane-associated Ubl4A is preferentially ubiquitinated. HEK293 cells were fractionated into an ER-containing membrane and cytosol fractions. Ubl4A immunoprecipitated from these fractions was analyzed by immunoblotting. (C) Ubl4A is conjugated with Lys48-linked ubiquitin chains. Ubl4A immunoprecipitated from extracts of cells expressing the indicated ubiquitin variants was analyzed by immunoblotting. (D and E) USP13 depletion causes accumulation of ubiquitinated Ubl4A in cells. (D) Ubiquitinated Ubl4A was analyzed in cells expressing the indicated shRNAs and HA-ubiquitin. (E) Ubiquitinated Ubl4A was analyzed in control wild-type cells and USP13 knockout (−/−) cells expressing HA-ubiquitin. (F) USP19 depletion does not affect Ubl4A ubiquitination. Asterisk indicates a non-specific band. (G) In vitro deubiquitination of Ubl4A by USP13. Ubl4A purified from USP13 knockout cells was treated with either buffer or increased concentration of FLAG-USP13 (0.15 µM–0.6 µM) at 37°C for 2 hr. Where indicated, Ub-aldehyde (Ub-Al, 1 µM) was included. (H) TCRα purified from USP13 knockout cells was incubated with either buffer or FLAG-USP13 (0.15 µM–0.6 µM) at 37°C for 2 hr.
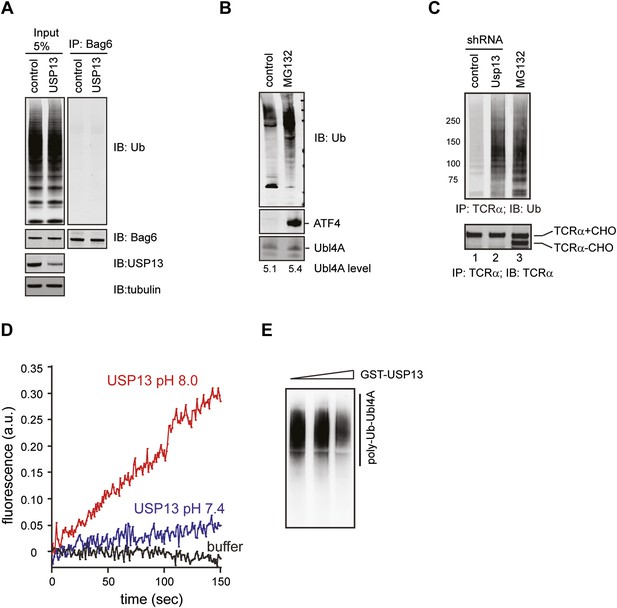
Regulation of Ubl4A by ubiquitination.
(A) USP13 does not regulate ubiquitination of Bag6. HEK293 cells were transfected with the indicated shRNA constructs. Endogenous Bag6 was immunoprecipitated under denaturing condition and was analyzed by immunoblotting. Where indicated, a fraction of the lysate (input) was analyzed directly by immunoblotting. (B) The proteasome inhibitor MG132 does not increase the steady state level of Ubl4A. Immunoblotting analysis of whole cell extracts from HEK293 cells either untreated or treated with MG132 (20 µM, 6 hr). The numbers show the quantification of Ubl4A levels. Note that MG132 stabilizes the transcription factor ATF4, but does not affect Ubl4A protein level. (C) Ubiquitinated TCRα accumulates in USP13 knockdown cells. (D) The USP13 activity is increased at pH 8.0. Purified FLAG-USP13 (120 nM) was incubated with Ub-AFC (2.5 µM) under the indicated pH condition. (E) Ubl4A purified from USP13 knockout cells was incubated with increased amount of GST-USP13 at 37°C for 2 hr.
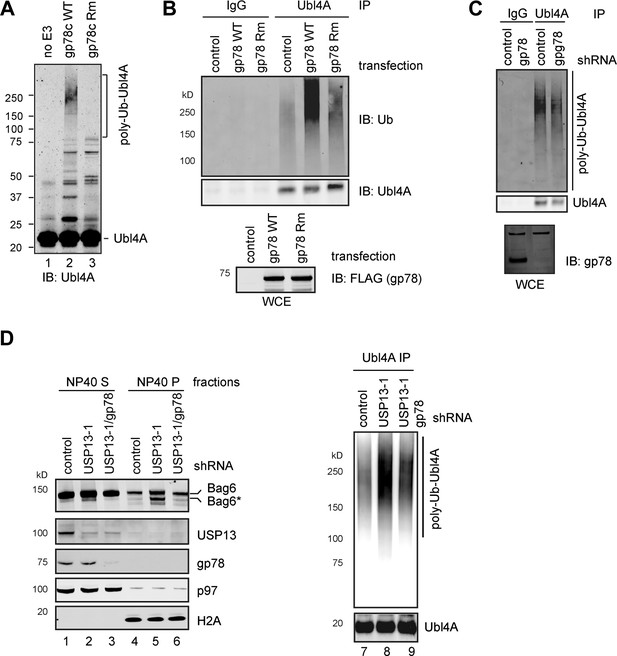
USP13 preferentially removes gp78-assembled ubiquitin chain from Ubl4A.
(A) In vitro ubiquitination of Ubl4A by gp78. Purified Ubl4A was incubated with E1, Ube2g2, ubiquitin, ATP in the absence (no E3) or presence of the wild-type gp78 cytosolic domain (gp78c) or a catalytically inactive gp78c RING mutant (Rm). The reactions were analyzed by immunoblotting with anti-Ubl4A antibody. (B) Overexpression of wild-type gp78 promotes ubiquitination of Ubl4A. HEK293 cells were transfected with control or the indicated gp78-expressing plasmid. Ubiquitination of endogenous Ubl4A was analyzed by immunoprecipitation followed by immunoblotting. WCE, whole cell extracts. (C) gp78 knockdown in cells does not significantly reduce basal ubiquitination of Ubl4A. Ubl4A immunoprecipitated under denaturing conditions from extracts of cells transfected with the indicated shRNA constructs was analyzed by immunoblotting. Where indicated, a fraction of the whole cell extract was directly analyzed by immunoblotting to verify the knockdown efficiency. Asterisk indicates a non-specific band, which serves as a loading control. (D) gp78 antagonizes USP13 in regulation of Ubl4A. Lane 1–6, A fraction of the cells expressing the indicated shRNAs and HA-Ubiquitin were subject to sequential extraction by NP40- and SDS-containing buffers. Lane 7–9, the remaining cells were used to measure the levels of ubiquitinated Ubl4A as in B.
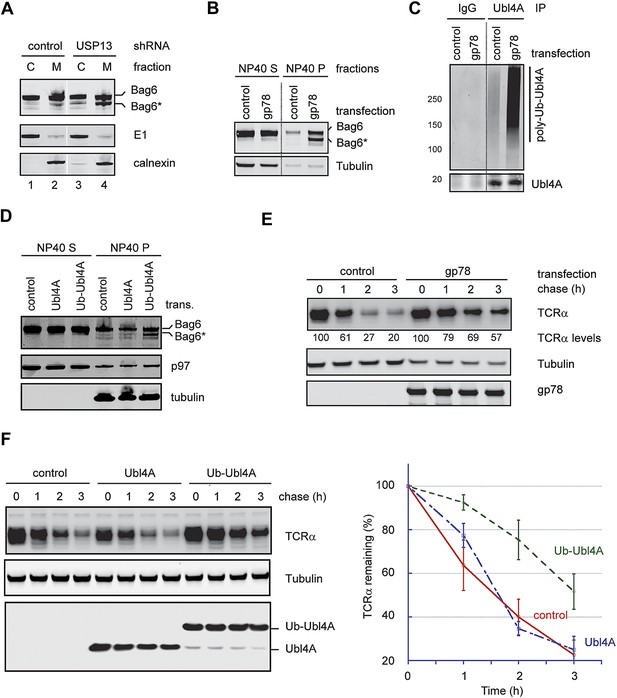
Hyper-ubiquitination of Ubl4A is associated with Bag6 clipping and ERAD inhibition.
(A) Membrane-associated Bag6 is preferentially cleaved in USP13 knockdown cells. C, cytosol fraction; M, membrane fraction. (B and C) Overexpression of gp78 causes cleavage of Bag6. (B) A fraction of the cells transfected as indicated were extracted sequentially with NP40- and SDS-containing buffers. The corresponding extracts were analyzed by immunoblotting. (C) Protein extracts from HEK293 cells transfected with control or gp78-expressing plasmid were subject to immunoprecipitation with Ubl4A antibodies or with IgG as a negative control. (D) Overexpression of a Ub-Ubl4A fusion protein induces Bag6 cleavage. As in B, except that cells expressing the indicated proteins were analyzed. (E) Overexpression of wild-type gp78 inhibits TCRα degradation. The number indicates the relative levels of TCRα averaged from two independent experiments. (F) Overexpression of Ub-Ubl4A inhibits TCRα degradation. The graph shows the quantification results from three independent experiments. Error bars, SD (n = 3).
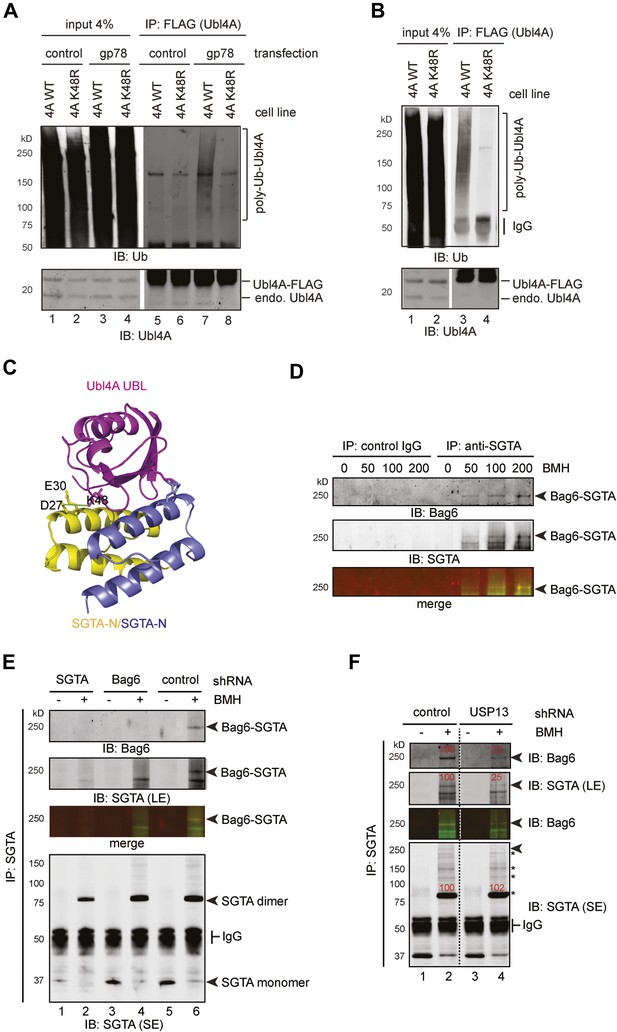
USP13 maintains a functional interaction between Bag6 and SGTA.
(A) Lys48 of Ubl4A is required for gp78-mediated ubiquitination in cells. Cells stably expressing wild type and K48R mutant Ubl4A were transfected with either an empty vector (control) or a gp78-expressing vector. Whole cell extracts were either directly analyzed by immunoblotting (lanes 1–4) or subject to immunoprecipitation with anti-FLAG antibodies prior to immunoblotting (lane 5–8). (B) Mutating Lys48 in Ubl4A reduces basal ubiquitination of Ubl4A. Ubl4A ubiquitination was analyzed using wild type and the K48R mutant Ubl4A cells transfected with an HA-Ub expressing plasmid. (C) A model of SGTA-N-Ubl4A UBL complex. The structure of SGTA-N domain (PDB: 4GOD) (Chartron et al., 2012) and Ubl4A UBL domain (PDB: 2DZI) was superimposed on the complex of the corresponding yeast complex (PDB: 2LXC) (Chartron et al., 2012). Note that Lys48 in Ubl4A interacts with two negatively-charged residues in SGTA. (D) A Bag6- and SGTA-containing complex can be crosslinked in cells. HEK293 cells were treated with the indicated concentrations of BMH prior to lysis. Cell extracts were subject to IP with either IgG as a control or anti-SGTA antibody followed by immunoblotting. The arrow indicates a crosslinking species that is detected by both anti-Bag6 and anti-SGTA blotting. (E) As in D, except that cells expressing the indicated shRNA constructs were used and that IP was only conducted with the anti-SGTA antibody. SE, short exposure; LE, long exposure. (F) As in E, except that cells transfected with either control or USP13 shRNA were used. Asterisks indicate SGTA-containing crosslinking products that are not affected by USP13 knockdown. The numbers show the relative levels for the crosslinking products.
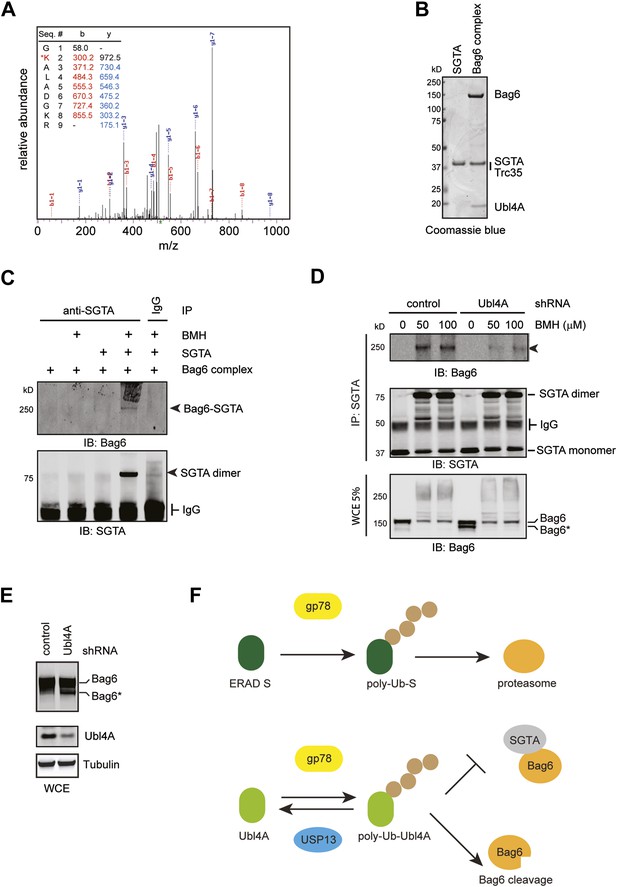
Bag6 and SGTA form a transient interaction dependent on Ubl4A.
(A) Mass spectrometry analysis of purified ubiquitinated Ubl4A. Purified Ubl4A was incubated with E2, Ube2g2, gp78, Ub, and ATP. After incubation, ubiquitinated Ubl4A was re-purified and analyzed by mass spectrometry. (B) Purified SGTA and the Bag6 ternary complex were analyzed by SDS-PAGE and Coomassie blue staining. (C) In vitro crosslinking demonstrates that SGTA can be crosslinked with the Bag6 complex. In vitro crosslinking was performed with the indicated proteins in the presence (+) or absence of 50 µM BMH. The reactions quenched by DTT were subject to immunoprecipitation with the indicated antibodies. The precipitated samples were analyzed by immunoblotting. (D) The formation of the 250 kD crosslinking product in cells requires Ubl4A. Control or Ubl4A knockdown cells were lysed under a denaturing condition. A fraction of the whole cell extracts (WCE), was analyzed by immunoblotting directly. The remaining samples were subject to immunoprecipitation with anti-SGTA antibody followed by immunoblotting analyses. The arrowhead indicates the 250 kD Bag6-SGTA crosslinking product, which is reduced under Ubl4A knockdown conditions. Note that a fraction of Bag6 also accumulated as the fast migrating Bag6* species in Ubl4A knockdown cells. (E) Ubl4A depletion also induces Bag6 cleavage in cells. Cells were directly lysed in the Laemmli buffer and the extracts were analyzed by immunoblotting. (F) USP13 helps sharpening the substrate specificity for gp78. gp78 can act not only on ERAD substrates but also on ERAD machinery proteins that interact with gp78 such as Ubl4A. USP13 antagonizes gp78 to remove ubiquitin conjugates from Ubl4A, thus prevent undesired ubiquitination by gp78.
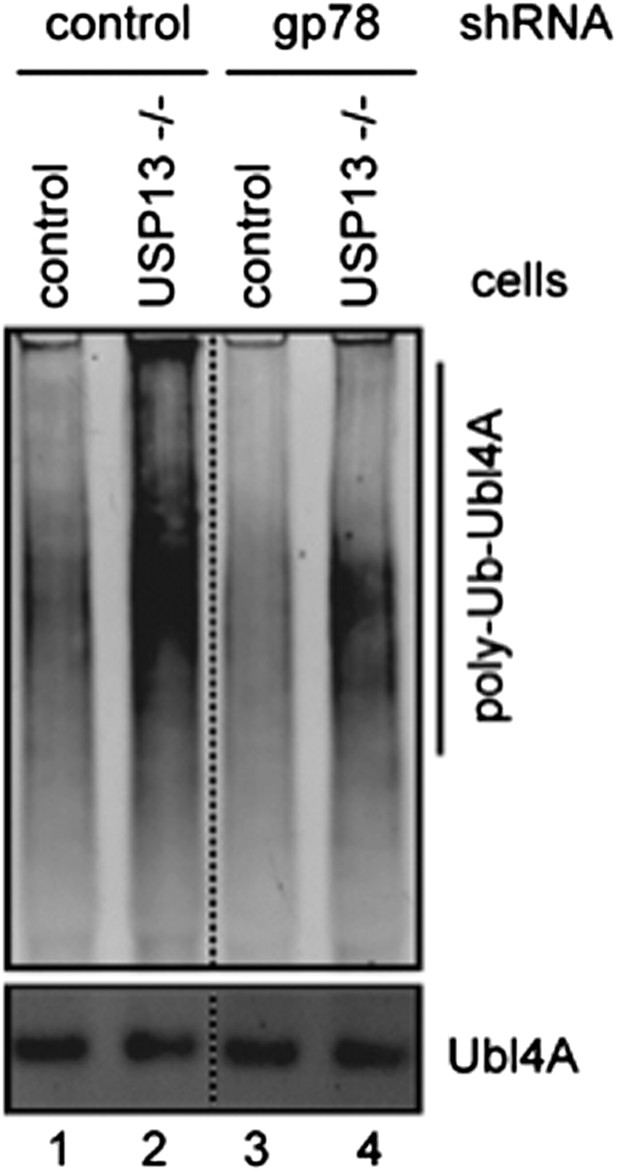
Author response image 1: Ubl4A ubiquitination was analyzed using control or USP13 knockout cells expressing either control or gp78 shRNA.
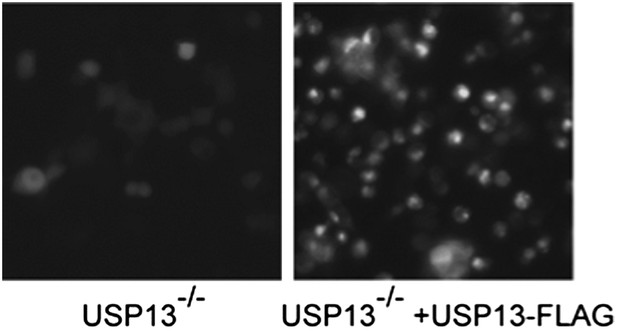
Author response image 2: USP13 knockout cells transfected with control or USP13 expressing plasmid together with TCRa-YFP were imaged using a Zeiss fluorescence microscope equipped with a 20X objective. The images were taken with the same exposure.
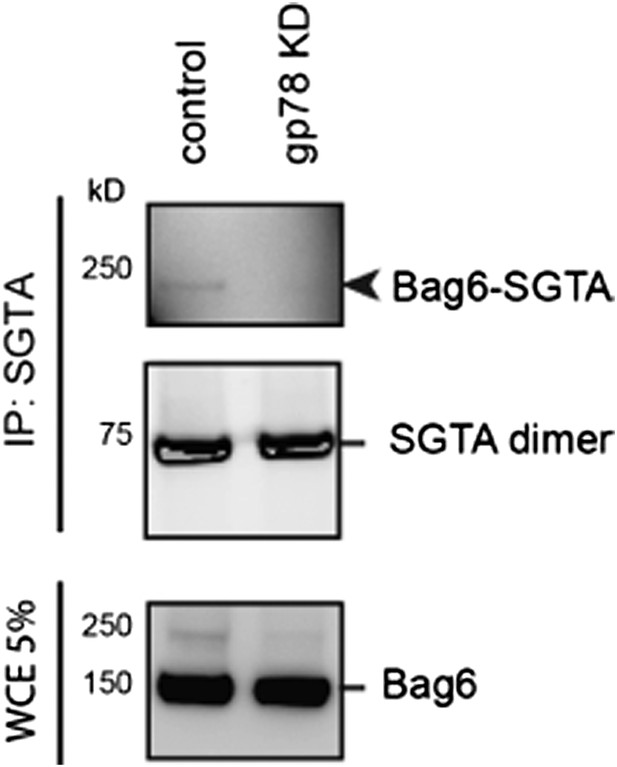
Author response image 3: Cells expressing either control or gp78 shRNA were treated with BMH. SGTA-containing complex was immunoprecipitated and analyzed by immunoblotting. A fraction of the whole cell extract was also analyzed. Note that the 250kD Bag6-SGTA crosslinking product was abolished in gp78 knockdown cells.





