Molecular insights into the origin of the Hox-TALE patterning system
Figures
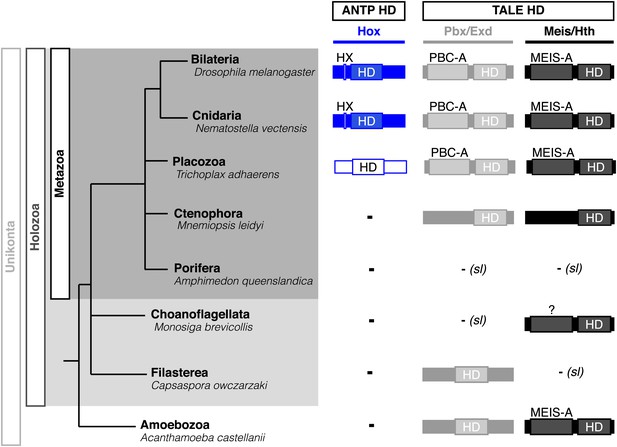
Phylogeny of Hox (ANTP superclass) and PBC/Meis (TALE superclass) proteins across eukaryote evolution.
Protein motifs required for Hox/PBC/Meis network are indicated when present: homeodomain (HD), hexapeptide (HX), PBC-A, and MEIS-A. Absence of the member in a given group is considered as resulting from a secondary loss (sl), when the ortholog is present more ancestrally. A question mark is indicated for Meis of Monosiga brevicollis because of incomplete sequence. The protein indicated in Placozoa is not coloured in blue since it is not a true Hox protein (see main text for details). Examples are provided for a representative species of each group. Members of PBC and Meis classes are called Pbx or Extradenticle (Exd) and Meis or Homothorax (Hth), respectively.
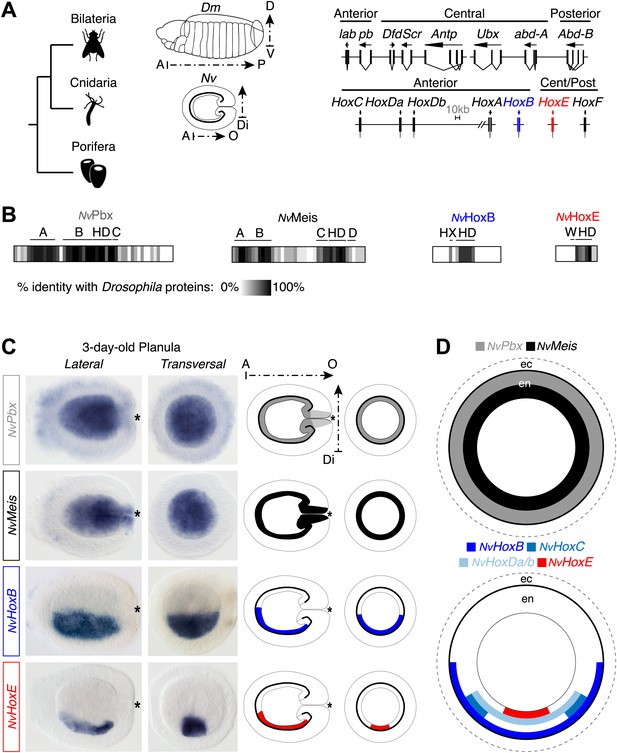
Hox and TALE members are co-expressed in the endoderm of the Nematostella embryo.
(A) Genomic organisation of Hox genes in Nematostella vectensis (Nv) and Drosophila melanogaster (Dm), two representative species of Cnidaria and Bilateria, respectively. Embryos at the planula stage are schematized; A–P: anterior–posterior; D–V: dorsal–ventral; A–O: aboral–oral; Di: directive axis. The Nematostella embryo is oriented according to recent findings (Sinigaglia et al., 2013). The nomenclature is calqued on (Chourrout et al., 2006) to avoid confusions with bilaterian Hox paralogs: NvHoxC (antHox7), NvHoxDa (antHox8), NvHoxDb (antHox8a), NvHoxA (antHox6), NvHoxB (antHox6a), NvHoxE (antHox1a), NvHoxF (antHox1). The two Nematostella Hox genes under study, NvHoxB and NvHoxE, are highlighted in blue and red respectively. Note that the same colour code is used in other figures. (B) Sequence identity between Nematostella and Drosophila proteins. NvHoxB and NvHoxE are compared to Labial (Lab) and Ultrabithorax (Ubx) respectively. The percentage of identity is represented by a grayscale gradient. Conserved domains in bilaterian TALE proteins are indicated (A, B, C, D). HX: hexapeptide. HD: homeodomain. See also Figure 2—figure supplements 1–4. (C) In situ hybridization of NvPbx, NvMeis, NvHoxB and NvHoxE in a three-day-old Nematostella planula. These four genes are expressed in the endoderm (en), as illustrated in (D). Ec: ectoderm. NvPbx and NvMeis are illustrated in grey and black, respectively. This colour code is used in other figures.
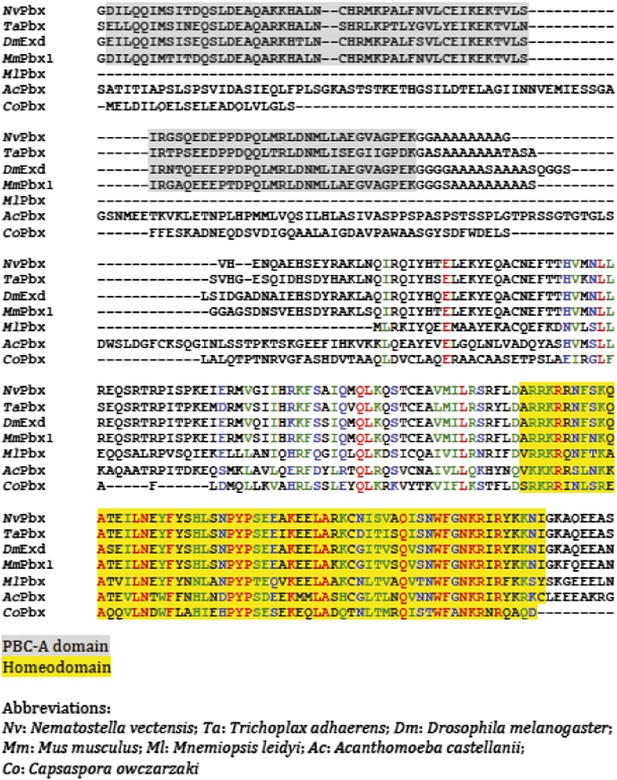
Protein sequence alignment of PBC members from representative species of Unikonta.
Regions encompassing the PBC-A domain and homeodomain (HD) are highlighted in grey and yellow respectively.
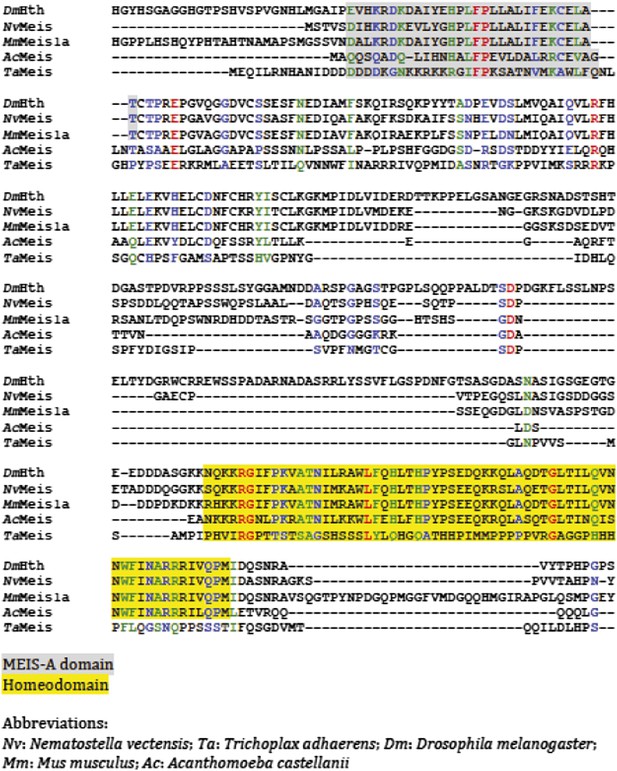
Protein sequence alignment of Meis members from representative species of Unikonta.
Regions encompassing the MEIS-A domain and homeodomain (HD) are highlighted in grey and yellow respectively.
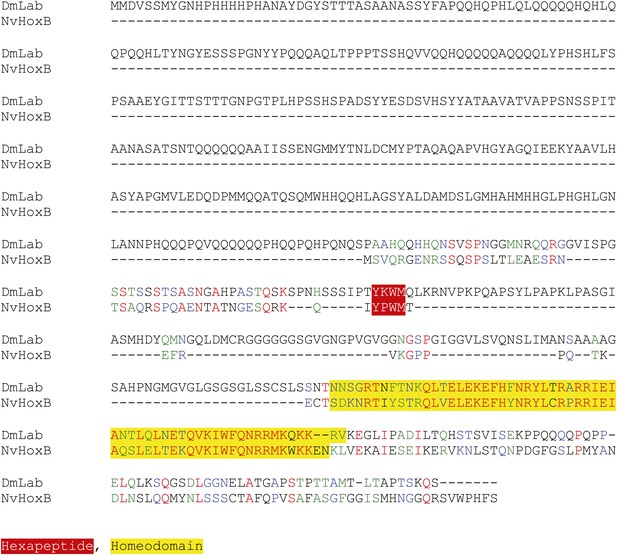
Protein sequence alignment between NvHoxB and the Labial (Lab) protein from Drosophila melanogaster.
Homeodomain and hexapeptide regions are highlighted in blue and red respectively.
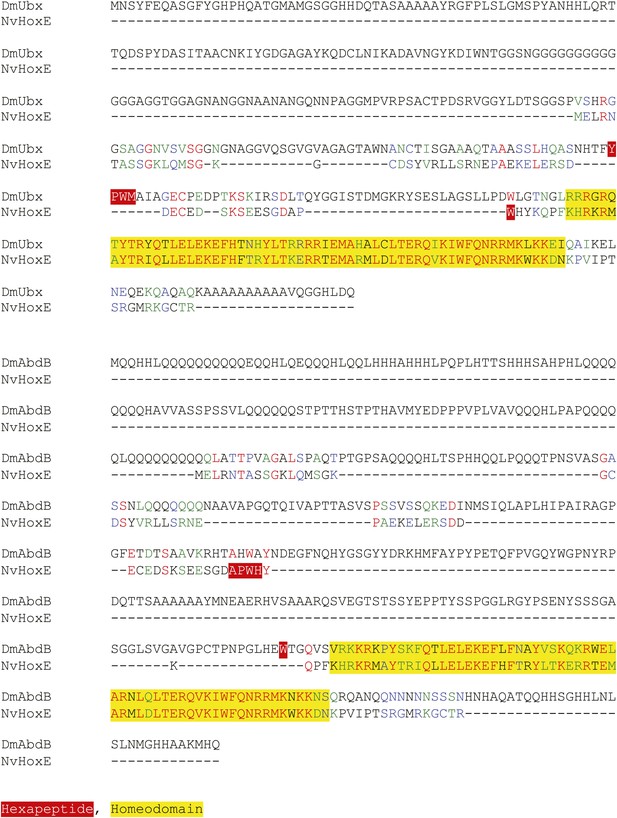
Protein sequence alignment between NvHoxE and the Ultrabithorax (Ubx) or AbdominalB (AbdB) protein from Drosophila melanogaster.
Homeodomain and hexapeptide regions are highlighted in blue and red respectively.
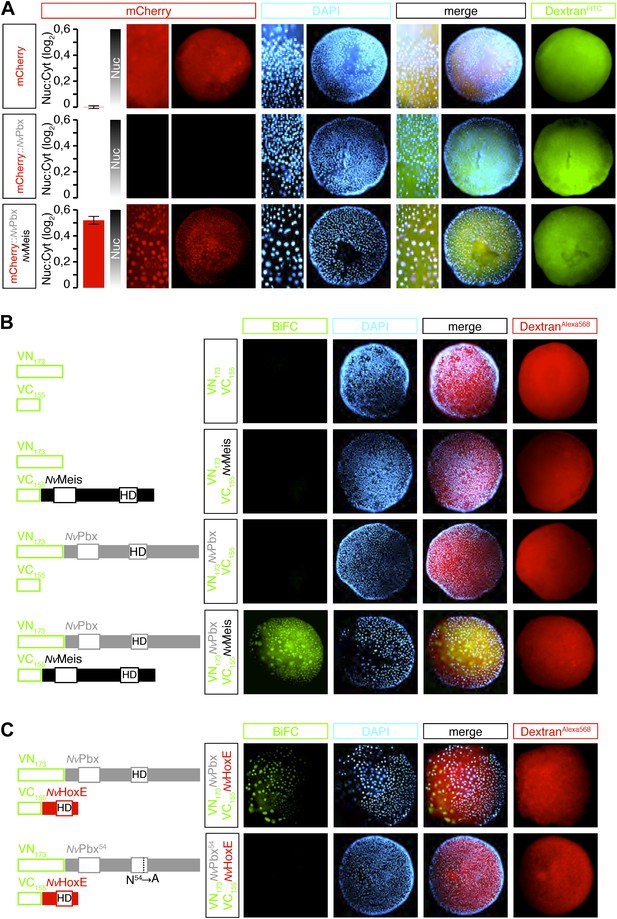
Hox and TALE members form protein complexes in vivo.
(A) NvPbx interacts with NvMeis in vivo. The nuclear localisation of a fusion mCherry-NvPbx protein was only observed upon co-injection with NvMeis. Graphs on the left are quantifications of the ratio between nuclear and cytoplasmic fluorescent signals (log2). Note that mCherry-NvPbx alone did not lead to any signal, suggesting that the fusion protein is not stable in the absence of Meis, as noticed in bilaterians. (B) BiFC between NvPbx and NvMeis in the Nematostella embryo. Fusion constructs are schematized on the left. VN: N-terminal fragment of Venus; VC: C-terminal fragment of Venus. Specificity of BiFC is verified by the absence of fluorescent signals upon the injection of isolated VN or VC fragments, together or with the complementary VC-NvMeis or VN-NvPbx fusion proteins, as indicated. Interaction between NvPbx and NvMeis occurs both in the cytoplasm and nucleus (see text for details). (C) BiFC between NvHoxE and NvPbx in the Nematostella embryo. Interaction occurs only in the nucleus. Mutation of the residue 54 in the homeodomain (HD) of NvPbx abolishes DNA-binding and BiFC with NvHoxE. In all panels, Dapi (cyan) stains nuclei and Dextran (red) is a control of injection conditions.
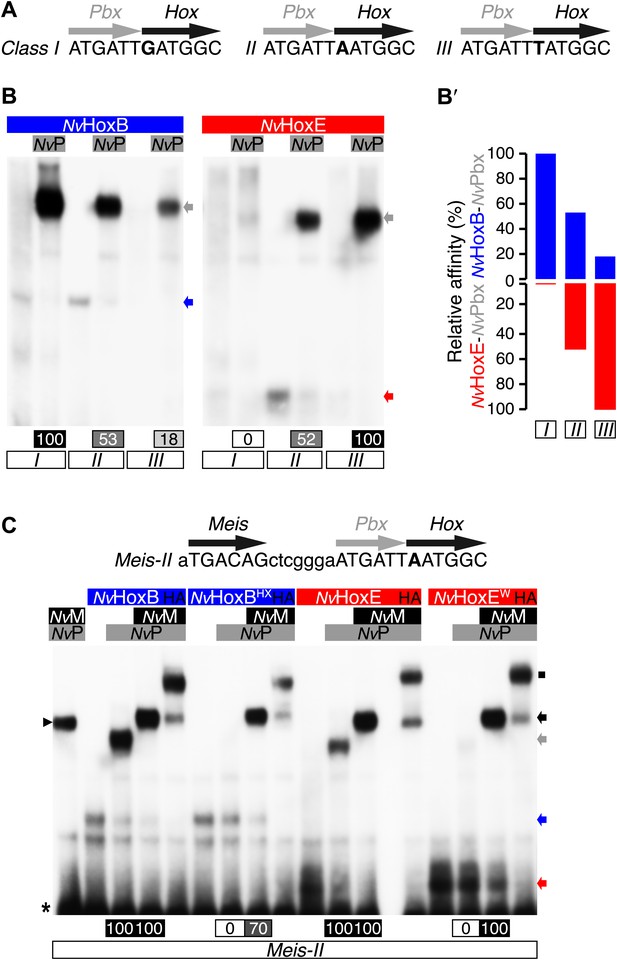
Interaction properties between NvHox and NvTALE proteins in vitro.
(A) Nucleotide sequence of the different classes of Hox/Pbx binding sites used in band shift experiments. The nucleotide that distinguishes each Hox/Pbx binding site is bolded. (B–B′) Band shift experiments between NvHoxB or NvHoxE and NvPbx on the three different classes of binding sites, as indicated. Coloured and grey arrows point to monomer or dimer binding, respectively. Graph on the right (B′) depicts the relative affinity of each dimeric complex on the three different binding sites, as deduced from the direct quantification on the gel (values are indicated at the bottom). (C) Band shift experiments between wild-type or HX-mutated NvHox proteins and NvTALE cofactors, as indicated. Colour codes and annotations are as in (B). Black arrow indicates trimeric NvHox/NvPbx/NvMeis complexes. Other bands are not specific (proteins of the lysate). Black scare highlights the supershift band resulting from the addition of an antibody against the HA tag of NvHox proteins. Asterisk shows the free probe. Note that the loss of dimeric NvHox/NvPbx complex upon the HX mutation is rescued in the presence of NvMeis.

Protein sequence alignment between NvHoxE (upper sequence) and NvHoxB (lower sequence).
The HX motif and HD are highlighted in red and yellow, respectively. A typical HX possesses a core tryptophan residue in a hydrophobic context, with a lysine or arginine residue at position +2 to +5 (In der Rieden et al., 2004).
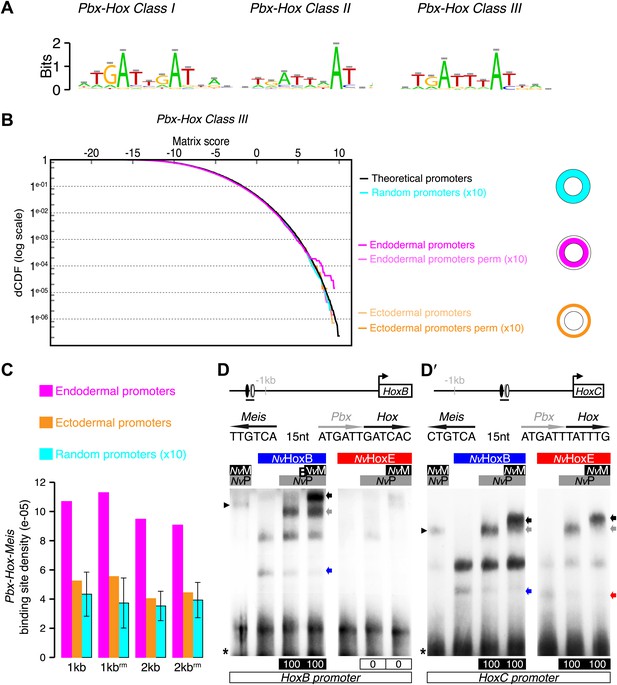
Genes expressed in the endoderm are enriched in Hox-TALE binding sites in their promoter region.
(A) Hox/Pbx binding motifs represented as logos. The three motifs represent the binding specificity of the Hox/Pbx complex for sites of class I, II, or III. Matrix was determined by Selex with the Drosophila proteins (Slattery et al., 2011). (B) Score distributions of the Hox/Pbx Class III matrix. The Y-axis is shown in logarithmic scale to highlight the relevant range of p values (small values). The separation of the pink curve (endoderm) from the black one (theoretical distribution) indicates an enrichment of the Hox/Pbx putative binding sites in the promoter of genes expressed in the endoderm. On the contrary, there is no enrichment in the promoter of genes expressed in the ectoderm, as the orange curve follows the black one. All negative controls also show no enrichment: random sets of gene promoters (cyan), promoter regions randomized by matrix column permutations for the endoderm (light pink) and ectoderm (light orange). (C) In silico analysis of Hox/Pbx/Meis binding sites in the promoter region (1 kb or 2 kbs upstream of the transcription start site) of genes expressed in the endoderm (pink), ectoderm (orange), or randomly chosen (cyan). The graph illustrates the preferential enrichment of Hox/Pbx/Meis binding sites in the promoter region of endodermal genes. Rm: repeat masked. (D–D′) Band shift experiments between NvHox and NvTALE proteins on binding sites found in the promoter region of NvHoxB and NvHoxC genes. Sequence and genomic position of each binding site are shown above the gel. Colour code and annotations are as in Figure 3. Note the distinct DNA-binding preferences of NvHoxB and NvHoxE on these two different target sites. See also Figure 5—figure supplements 1 and 2.
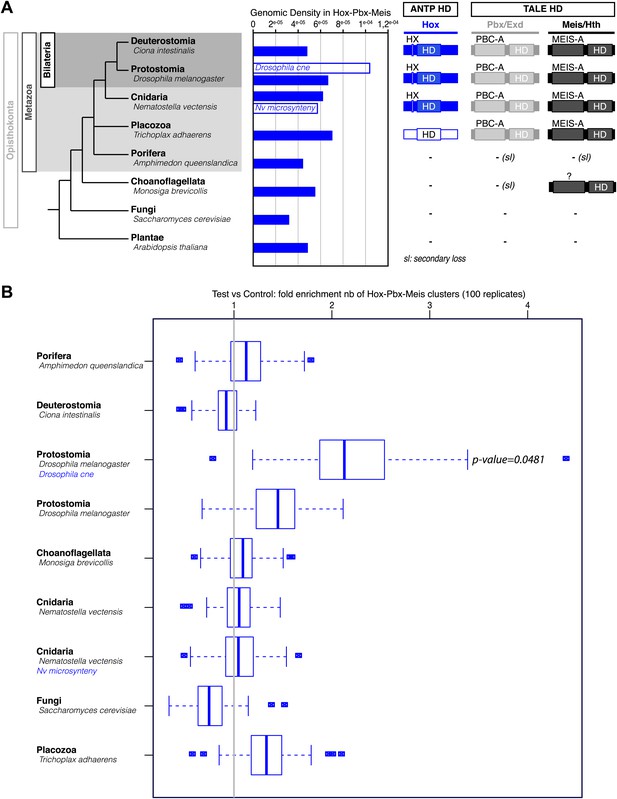
Genome-wide analysis cannot reveal significant enrichment of Hox/PBC/Meis binding sites.
(A) Density of Hox/PBC/Meis clusters in the genome of various organisms. The density was calculated on all non-coding regions of the genomes (blue-filled bars), repeat-masked except for Amphimedon where the repeat-masked genome was the same as the non-repeat-masked one. Two other conditions (white bars) represent a subset of the genome. The non-coding regions (CNE) conserved in 12 Drosophila genomes show a higher density than genome-wide approaches. These results suggest that the search space needs to be reduced to obtain a good signal/noise ratio. One idea would be to search for conserved regions within cnidarians, taking Nematostella as reference, but such an analysis was beyond the scope of this project, and the divergence time of available cnidarian genomes might not be adapted to this analysis. We attempted a related analysis by taking microsyntenic regions as described in Irimia et al. (2012). The results do not show a high density comparable to the Drosophila CNEs. We hypothesize that these microsyntenic regions still have a low signal to noise ratio for these motif clusters, or that biologically, the Hox/Pbx + Meis cluster is not majorly involved in the regulation taking place at these regions conserved throughout metazoans. (B) Validation of cluster enrichment. For each organism, we calculated the enrichment of Hox/PBC/Meis clusters, compared to a random control. At first, we conducted this analysis on random sequences, artificially generated from Markov models trained on the genomes of interest, and taking into account the number and positions of the repeats. The results, however, were highly dependent on the order of the Markov model. To circumvent this, we used randomized motifs as a control, allowing us to keep working on the real genomic sequences. We permuted the motif positions 100 times, allowing, removing the biological signal, while retaining the statistical properties of the PSSMs describing the motifs. We observed a clear enrichment of the clusters in Drosophila CNEs. In contrast, most of the genome-wide analyses did not reveal any enrichment, except in Drosophila, which showed a slight enrichment similar to Trichoplax. As Trichoplax has no HX-containing ANTP proteins (therefore no Hox/PBC/Meis network), with signal to noise ratio still quite low, we did not consider this slight enrichment.
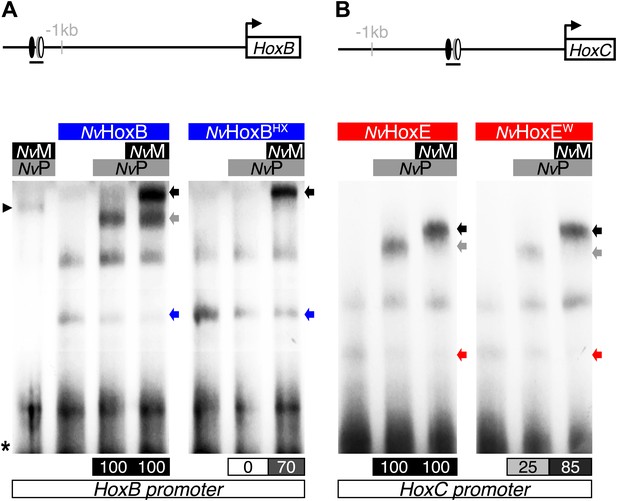
NvMeis promotes HX-independent interaction modes on DNA-binding sites found in the promoter region of NvHoxB (A) and NvHoxC (B).
Band shift experiments are performed with wild-type or HX-mutated forms of NvHoxB and NvHoxE, in the presence of NvTALE cofactors, as indicated. Loss of dimeric NvHox/NvPbx complexes upon the HX mutation is rescued in the presence of NvMeis. Values of the interaction levels are shown at the bottom of the gel. Asterisk marks the free probe. Black arrowhead (left) corresponds to the dimeric NvPbx/NvMeis complex. Coloured, grey, and black arrows (right) show monomer, dimer, and trimer DNA-binding complexes, respectively.
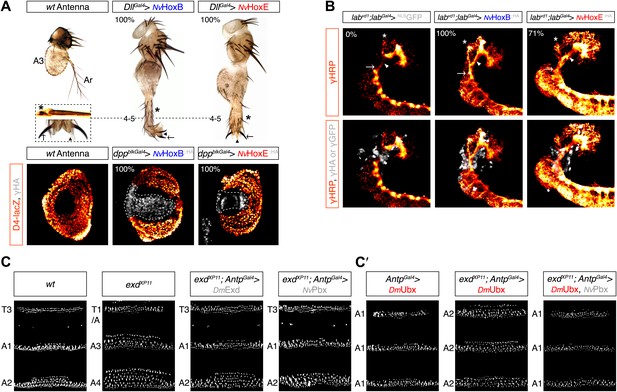
Functional analysis of NvHox and NvPbx proteins in Drosophila.
(A) Antenna-to-leg transforming activities of NvHoxB and NvHoxE. NvHox proteins were expressed in the antenna with the Distalless (Dll)-Gal4 driver. Asterisk depicts leg-specific bracted bristles. 4–5 shows the transformation of the arista in two tarsal segments. Arrow and arrowhead in the enlargement indicate the formation of the leg-specific terminal claw and its associated sensory pad respectively. The antenna-to-leg transformation by NvHox proteins (grey) is achieved through the repression of the spineless (ss) target gene, as observed by the repression of the ss enhancer D4 activity on lacZ reporter gene expression (orange). See also Figure 6—figure supplements 1 and 2. (B) Rescue of the labial (lab) mutant phenotype in the tritocerebrum by NvHox proteins. The central nervous system is stained with an anti-HRP (orange). Hox or GFP (as a control) proteins (grey) are expressed in the tritocerebrum with a lab-Gal4 driver. Frontal connectives (asterisk), longitudinal connectives (arrowhead) and tritocerebral commissure (arrow) are indicated. In lab mutant background, longitudinal connectives are reduced, frontal connectives project ectopically and the tritocerebral commissure is missing (Hirth et al., 1998). Expression of NvHoxB or NvHoxE in this mutant context leads to a complete or strong rescue of this phenotype, respectively. See also Figure 6—figure supplement 2. (C–C′) NvPbx can rescue zygotic exd mutant phenotypes in the Drosophila larva cuticle. (C) Larvae homozygous for the zygotic exdXP11 mutation have T3 and A1 segments that resemble to a T1/abdominal or A3 segment, respectively. Thoracic expression of either DmExd or NvPbx in this mutant background (through the UAS/Gal4 system, with the Antennapedia (Antp)-Gal4 driver) is sufficient to restore the correct specification of T3 and A1, as assessed by the shape and arrangement of denticle belts. (C′) Ubx normally specifies the A1 segment. Ectopic expression of Ubx with Antp-Gal4 induces A1-like segments in the thorax. In absence of Exd, Ubx produces A2-like segments. Providing back NvPbx in this genetic background is sufficient to restore the normal A1-inducing activity of Ubx. See also Figure 6—figure supplements 3 and 4.
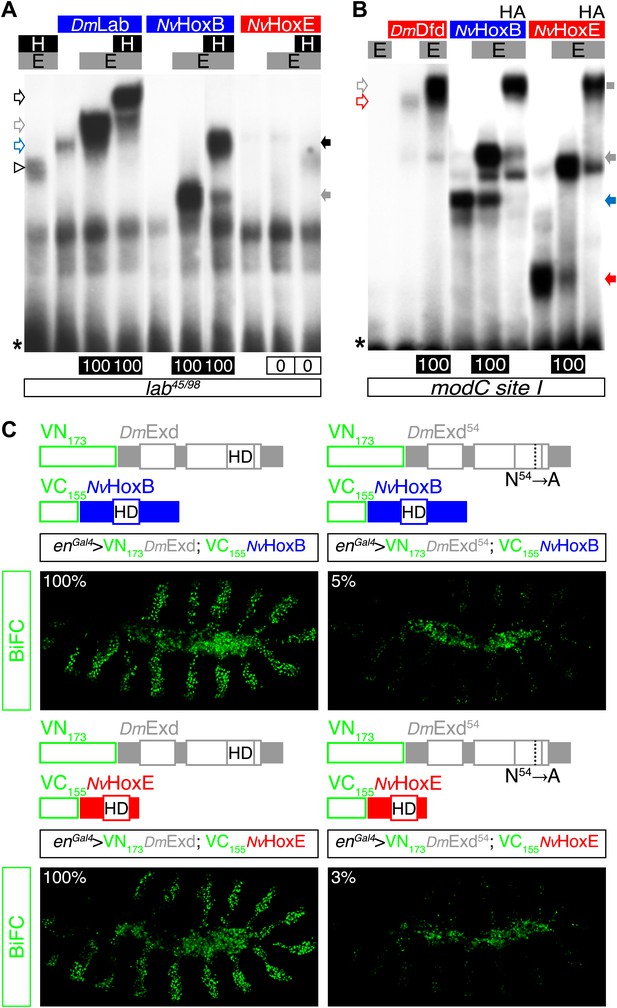
NvHoxB and NvHoxE interact with the Drosophila TALE cofactors Extradenticle (Exd) and Homothorax (Hth) in vitro and in vivo.
(A and B) Band shift experiments on physiological target sequences of Drosophila Hox proteins. Probes derived from the auto-regulatory element of the Hox genes labial (lab48/95, A) and Deformed (Dfd, modCsite 1, B), corresponding to binding sites of class I or II, respectively. Colour codes and annotations are as in Figure 5—figure supplement 2. Unfilled arrows indicate DNA-binding complexes with Drosophila Hox (DmLab and DmDfd) and TALE (E and H) proteins. Grey scare in (B) indicates the supershift band resulting from the addition of an antibody against the HA tag of NvHox proteins. (C) Visualisation of interactions between NvHox proteins and Exd by BiFC in a live stage 10 Drosophila embryo. Fusion constructs are schematized above each picture. Constructs were expressed with the engrailed (en)-Gal4 driver. Specificity of BiFC was verified with the Exd fusion construct mutated in the residue 54 of the HD.
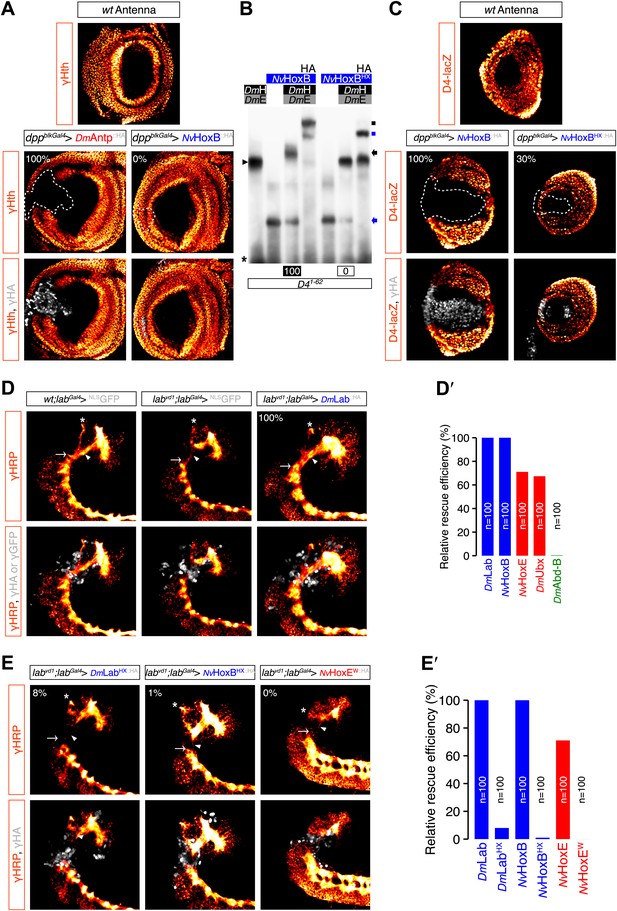
Role of the HX of NvHox proteins in generic Drosophila Hox assays.
(A) Antenna-to-leg transforming activities of NvHox proteins do not rely on the repression of homothorax (hth) target gene. Expression of NvHoxB (grey) in the proximal part of the antenna imaginal disc has no effect on hth expression (as assessed by an immunostaining against Hth, orange). By comparison, expressing the Drosophila Antennapedia (Antp) in the same condition leads to a complete loss of Hth. (B) The HX mutation abolishes complex formation between NvHoxB and Drosophila TALE cofactors on the target binding sites of the spineless D4 regulatory element. Colour codes and annotations are as in Figure 5—figure supplement 2. (C) The HX mutation affects the repressive activity of NvHoxB on the spineless D4 target element in vivo. Proteins were expressed as in (A). Expression of D4 was indirectly quantified through immunostaining against the D4-driven LacZ reporter protein (orange). (D–E′) The HX mutation abolishes the rescue activity of Drosophila and Nematostella Hox proteins in the tritocerebrum of labial (lab) mutant embryos. (D–D′) Rescue assays with wild-type Hox proteins, as indicated. (E–E′) Rescue assays with HX-mutated Hox proteins, as indicated. The central nervous system is stained with an anti-HRP (orange). HA-tagged proteins (grey) were expressed in the tritocerebrum with a lab-Gal4 driver. For quantifications of rescue efficiency (D′–E′), 100 embryos were examined in each case. Percentage values were corrected in order to take into account the phenotypic penetrance of the lab mutation in tritocerebral development (90%). The rescue efficiency of Dmlab was used as a standard and taken as 100%. The rescue values (the relative percentage of embryos showing a complete rescue of the tritocerebral brain defects) of DmHox and NvHox gene products are shown in percentage relative to Dmlab.
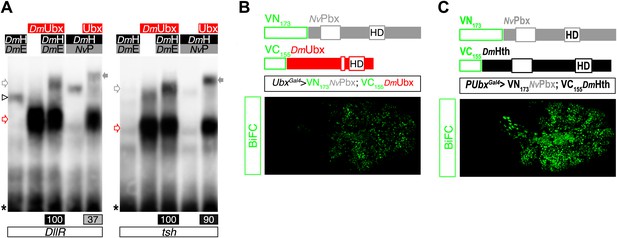
NvPbx interacts with the Drosophila Ultrabithorax (Ubx) and Homothorax (Hth) proteins in vitro and in vivo.
(A) Band shift experiments on DNA-binding sites derived from the regulatory elements of the Distalless (DllR) and teashirt (tsh) Hox target genes. Colour code and annotations are as in previous figures. (B–C) NvPbx interacts with Ubx (B) and Hth (C) in vivo. Fusion proteins generated for BiFC are schematized above the pictures. Expression was conducted with the Ubx-Gal4 driver. Pictures were acquired in a live stage 10 embryo.
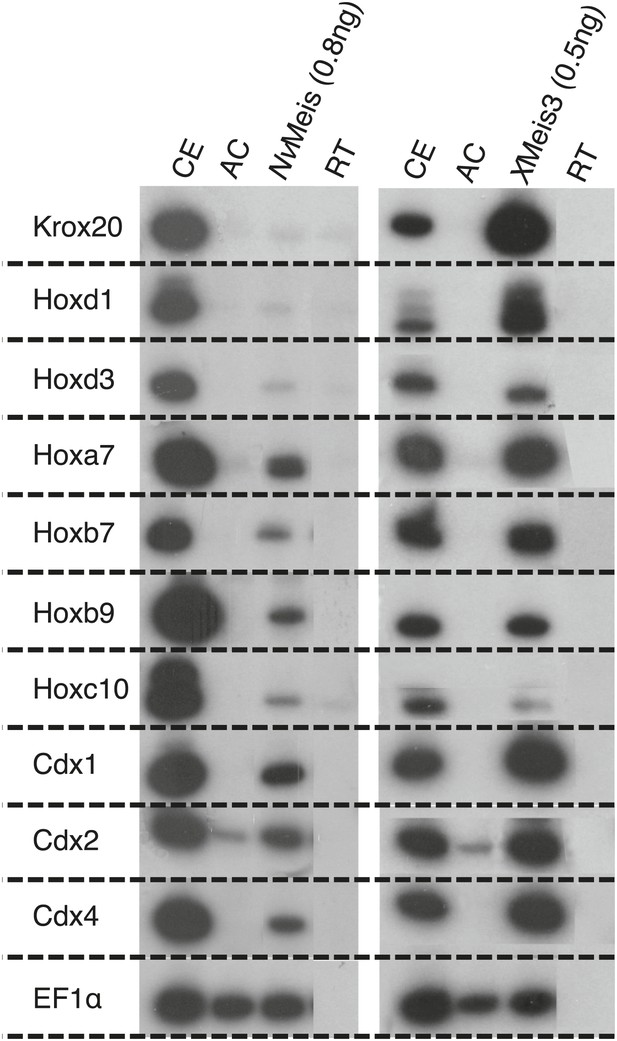
NvMeis reproduces generic bilaterian Meis activities in the Xenopus embryo.
While more anterior hindbrain marker genes are not expressed (krox20, hoxd1 and hoxd3), more posterior spinal cord marker genes such as, hoxa7, hoxb7, hoxb9, hoxc10, hoxd10, cdx1, cdx2 and cdx4 are robustly induced upon injection of NvMeis in the embryo. Total RNA was isolated from five control embryos (CE) and eighteen animal caps (AC) from the control or injected groups. Semi-quantitative RT-PCR analysis was performed on AC explants that were removed from control (non injected, RT) and injected embryos cultured until the neurula stage 16. EF1αcontrols for RNA levels in each sample. A comparative level of genes induction by XMeis3 is provided on the right.
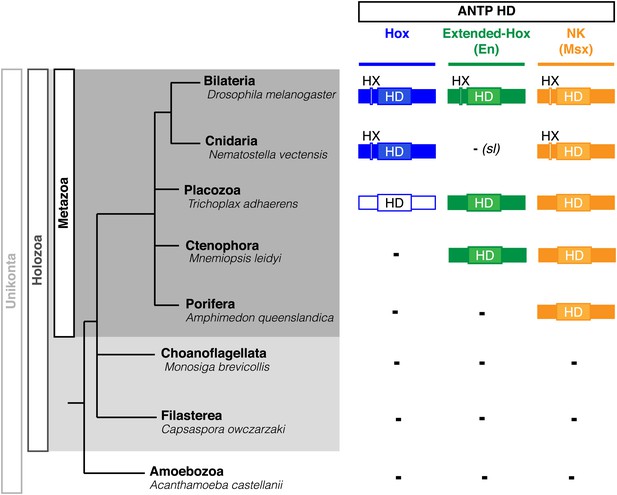
Phylogeny of Hox, Extended-Hox (Engrailed, En) and NK (Msx) proteins during eukaryote evolution.
Nomenclature is as in Figure 1. The conserved features of any HX motif correspond to a sequence containing an invariant tryptophan residue in a hydrophobic context, with a lysine or arginine residue at position +2 to +5, as previously defined (In der Rieden et al., 2004).
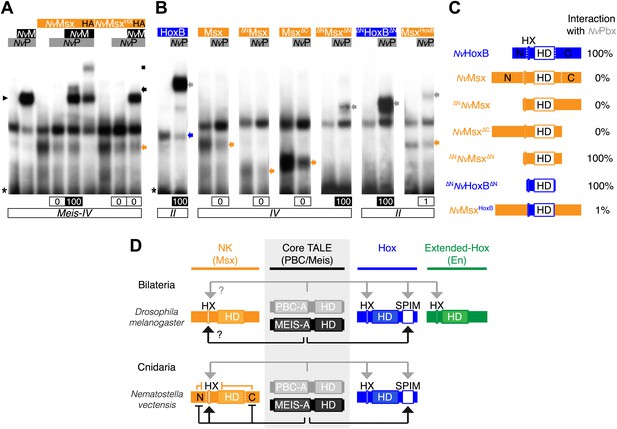
Genesis of Hox–TALE interaction networks in Metazoa.
(A) Band shift experiments between NvMsx and NvTALE cofactors, as indicated. Colour code and annotations are as in previous figures. Note that no dimeric NvMsx/NvPbx complex is formed. Binding reactions with NvMsx proteins were performed on a consensus Msx/Pbx binding site derived from vertebrates and containing an additional Meis binding site in a topology similar to the Hox probe (‘Materials and methods’). (B) Band shift experiments between wild-type, truncated or chimeric NvMsx and/or NvHoxB proteins, and NvPbx, as indicated. (C) Scheme of the diverse protein constructs and their corresponding interaction affinity level with NvPbx, as assessed from quantification of each band shift. Quantifications with truncated NvMsx proteins were deduced by comparison with the trimeric NvMsx/NvPbx/NvMeis in (A). (D) Molecular rules underlying interaction properties between NK, Hox, or extended-Hox members and TALE cofactors in Eumetazoans. In this model, Meis is able to promote HX-dependent interactions between NvMsx and NvPbx by masking inhibitory interaction domains in NvMsx. Whether a similar role could exist in Bilateria remains to be determined. We noticed that the Drosophila Msx protein contains an HX and forms trimeric but not dimeric complexes with the Drosophila TALE partners (not shown). HX-dependency in those interactions remains to be determined (question marks). Interaction between DmEn and PBC is also HX-dependent but does not require the presence of Meis to occur. In contrast to the NK or extended-Hox families, most members of the Hox family have retained a HX motif. This motif is required for generic Hox/Pbx functions. The additional presence of Meis allows revealing specific Pbx interaction motifs (SPIMs), which could be important for distinguishing and/or diversifying the embryonic activities of each Hox paralog group member. See also Figure 8—figure supplement 1.
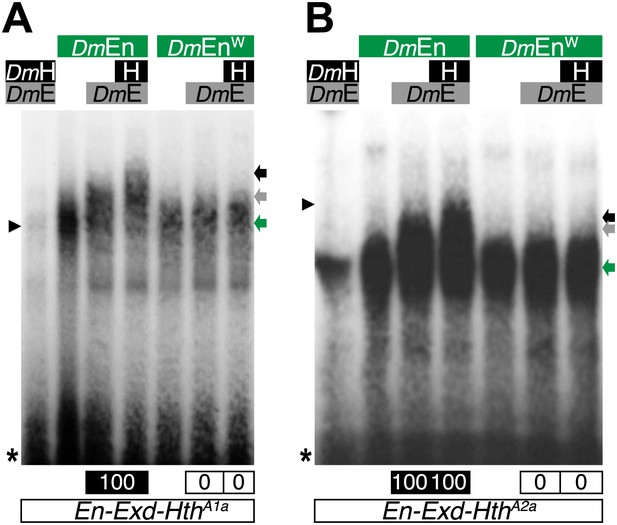
The Drosophila Engrailed (DmEn) protein forms HX-dependent DNA-binding complexes with Exd (DmE) and Hth (H) on physiological target sites.
Colour code is as in previous figures. Monomer binding of DmEn is indicated by a green arrow.
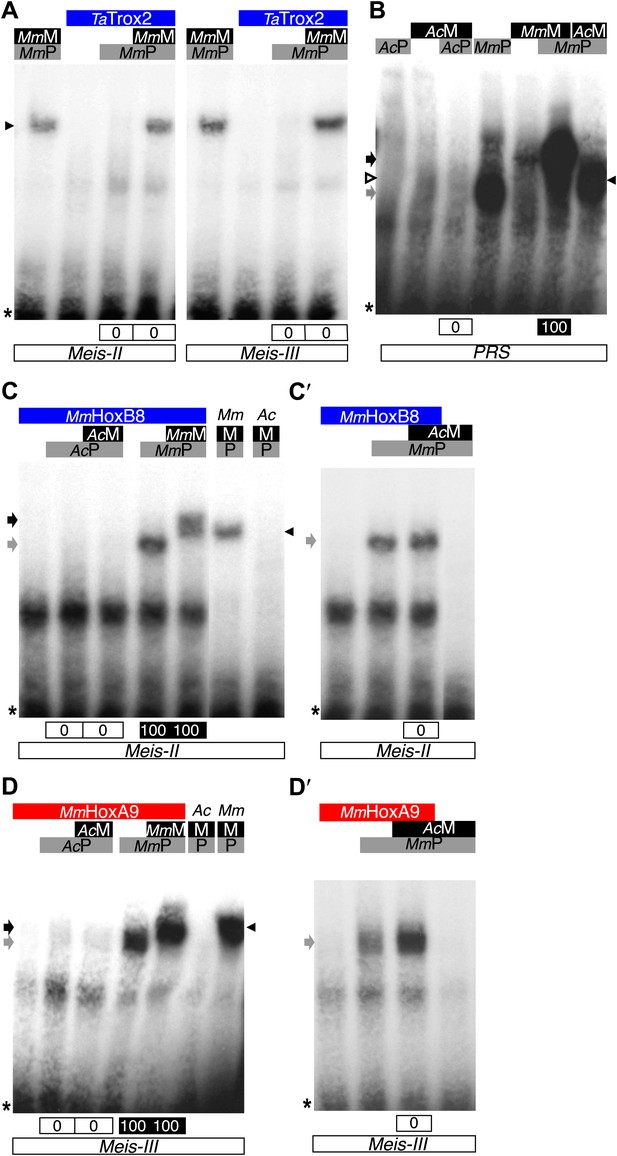
Interaction properties of Hox and TALE proteins from Trichoplax adhaerens and Acanthamoeba castellanii.
(A) The ProtoHox/ParaHox Trox2 protein from Trichoplax adhaerens does not form DNA-binding complexes with PBC or PBC/Meis in vitro. Band shift experiments are performed with mouse Pbx (MmP) and Meis (MmM) proteins on central (Meis-II) and posterior (Meis-III) Hox/PBC/Meis binding sites as indicated. Black arrowhead shows dimeric Pbx/Meis complexes. (B–D′) PBC (AcP) and Meis (AcM) proteins from the unicellular Acanthamoeba castellanii organism cannot form protein complexes between each other or with Hox proteins. (B) Band shift experiment on a consensus PBC binding site (PRS, Chang et al., 1995). AcP does not bind DNA, neither as a monomer nor with AcM. A weak monomer DNA-binding of AcM is observed (white arrowhead). This monomer binding is strongly enhanced in the presence of mouse Pbx1 (MmP, black arrowhead). In comparison, MmP binds strongly (grey arrow), and the monomer binding of Meis1 (MmM) is also strongly enhanced in the presence of Pbx1 (black arrow). (C–C′) Band shift experiments with mouse HoxB8 (MmHoxB8) and mouse or Acanthamoeba TALE cofactors on the central (Meis-II) Hox consensus binding probe as indicated. (D–D′) Band shift experiments with mouse HoxA9 (MmHoxA9) and mouse or Acanthamoeba TALE cofactors on the posterior (Meis-III) Hox consensus binding probe as indicated. Complexes with Hox proteins are observed only with mouse Pbx (grey arrows) and Pbx/Meis partners (black arrows) on both probes. AcPbx and AcMeis proteins are not able to form dimeric complexes on these probes, unlike mouse TALE proteins (black arrowheads). See also Figure 9—figure supplement 1.
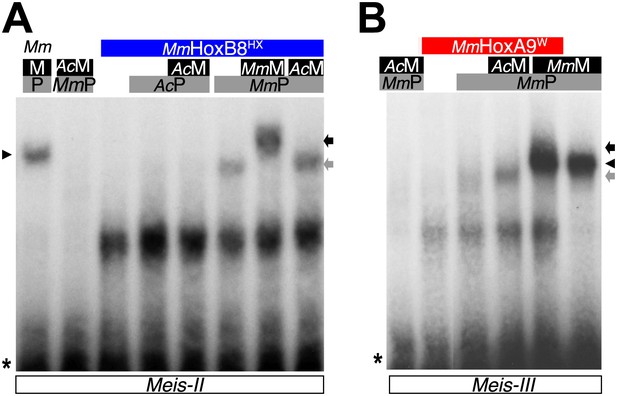
PBC and Meis proteins from the unicellular Acanthamoeba castellanii organism do not form protein complexes with mouse HX-mutated Hox proteins.
(A) Band shift experiment with HX-mutated HoxB8 on a central consensus binding site (Meis II). (B) Band shift experiment with HX-mutated HoxA9 on a posterior consensus binding site (Meis III). Colour code and annotations are as in previous figures. Protein complexes are only observed with mouse proteins, as previously described (Hudry et al., 2012).
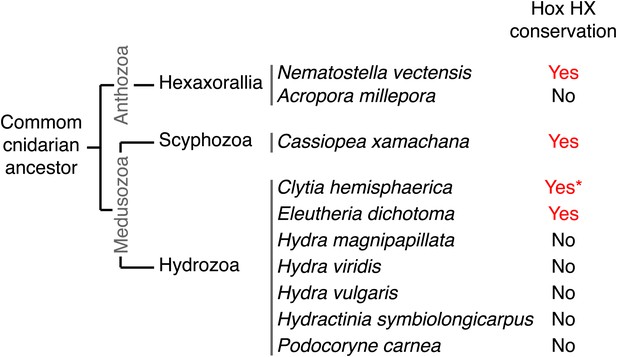
Phylogeny of the HX in Hox proteins of main cnidarian lineages.
https://doi.org/10.7554/eLife.01939.025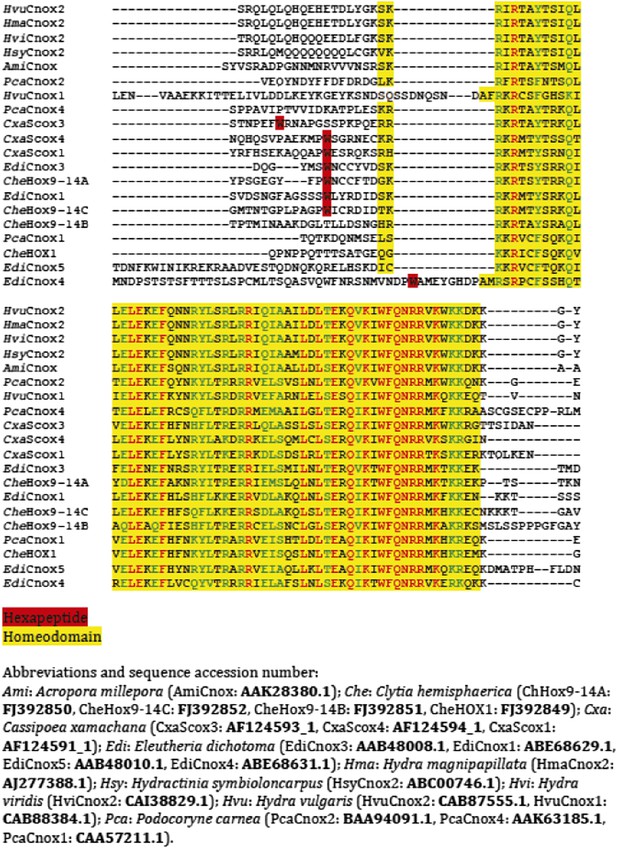
Protein sequence alignment of the region encompassing the HX (highlighted in red) and HD (highlighted in yellow) of cnidarian Hox members used in Figure 10—figure supplement 1.
https://doi.org/10.7554/eLife.01939.026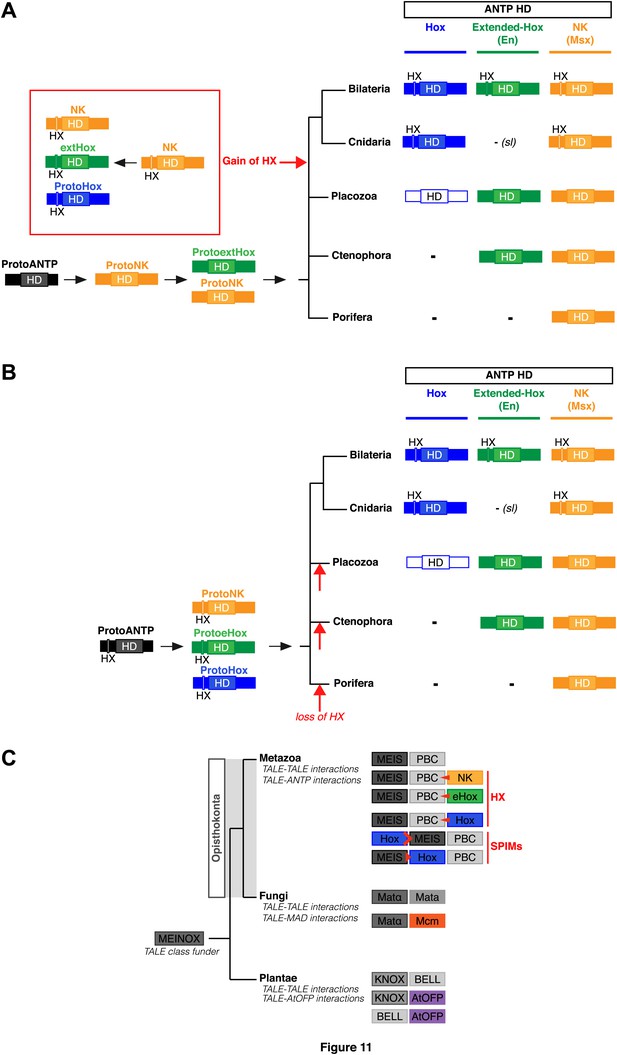
Evolution of molecular signatures underlying interaction networks with TALE proteins.
(A) Model for the apparition of the HX in ANTP members of Cnidaria and Bilateria. In this scenario, the HX appeared in a duplicated NK protein that gave rise to other HX-containing ANTP members in the common ancestor of Bilateria and Cnidaria. Duplications in other lineages occurred without the apparition of the HX. (B) Second model for the apparition of the HX in ANTP members of Cnidaria and Bilateria. This model is based on a recent work postulating the existence of Hox/ParaHox, NK, and extended-Hox clusters in the common ancestor of animals (Mendivil Ramos et al., 2012). In this scenario, it is unlikely that the HX appeared after duplications of the ProtoANTP ancestor independently and at the same place in the ProtoHox, ProtoNK and ProtoExtended(e)Hox families. Thus, the HX motif was probably already present in the ProtoANTP ancestor. This motif was secondarily lost in ANTP members of Porifera, Placozoa and Ctenophora during evolution. See also ‘Discussion’. (C) TALE proteins and the generation of interaction networks across major multicellular branches of the eukaryote evolutionary tree. TALE proteins are indicated in grey-filled boxes, with different grey tones corresponding to different TALE members. Other colours depict non-TALE members. Interactions can occur between different TALE members, or between TALE and non-TALE members. These interactions involve different proteins in each major multicellular branch, as indicated. Red signs in Metazoa symbolise interaction modes involving the HX or specific PBC interaction motifs (SPIMs) between TALE (PBC/Meis) and ANTP (NK, extended(e)Hox, Hox) members.
Additional files
-
Supplementary file 1
Constructs generated for BiFC, functional and shift assays.
- https://doi.org/10.7554/eLife.01939.028





