DNA polymerase V activity is autoregulated by a novel intrinsic DNA-dependent ATPase
Figures
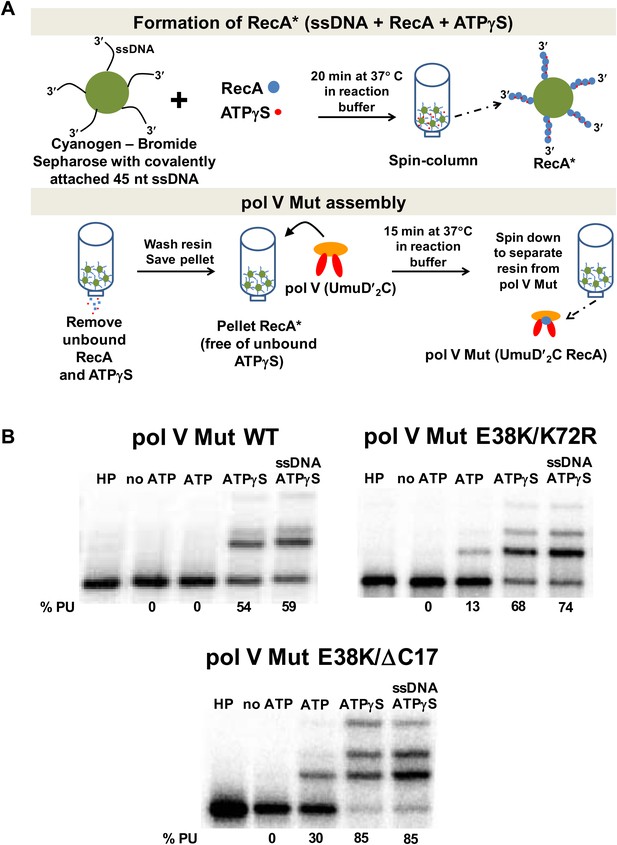
Pol V Mut requires ATP/ATPγS for activity.
Sketch of pol V Mut assembly; pol V is activated by RecA* bound to Cyanogen-Bromide Sepharose resin as described in ‘Materials and methods’. The pol V Mut assembly protocol ensures the separation of pol V Mut from free RecA, ssDNA and ATPγS. (B) Pol V Mut (400 nM) activity was detected on 5′-32P-labeled 3 nt oh HP (100 nM) in the presence or absence of ATP/ATPγS and dNTPs. To detect free RecA in the pol V Mut solution, ssDNA and ATPγS was added to the reaction. Comparable activity levels between ATPγS alone and ATPγS + ssDNA indicate that pol V Mut is intact and free of RecA.
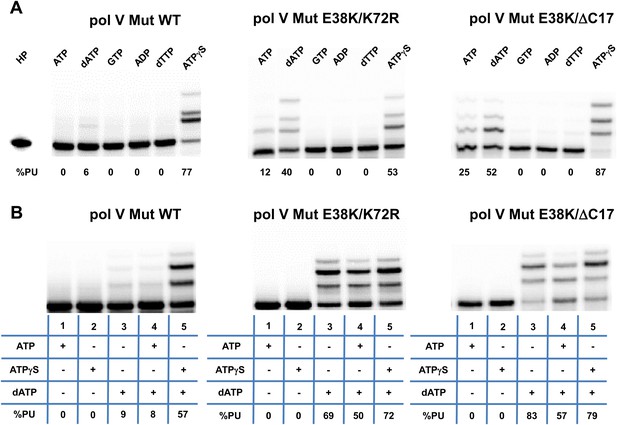
Pol V Mut is not activated by GTP, ADP, or dTTP and does not incorporate ATP/ATPγS in to DNA during synthesis.
Pol V Mut WT, pol V Mut E38K/K72R, and pol V Mut E38K/ΔC17 were assembled according to the protocol in Figure 1A. (A) ATP, dATP, GTP, ADP, dTTP or ATPγS (500 μM) were used to activate pol V Mut for DNA synthesis and activity was checked in the presence of dNTPs (500 μM) and 3 nt oh HP (50 nM). (B) A HP containing TTT as its 3 nt oh was employed to determine if the various pol V Muts insert ATP or ATPγS during DNA synthesis. DNA extension is only observed in reactions where dATP is included. Other dNTPs are not present in any of the reactions.
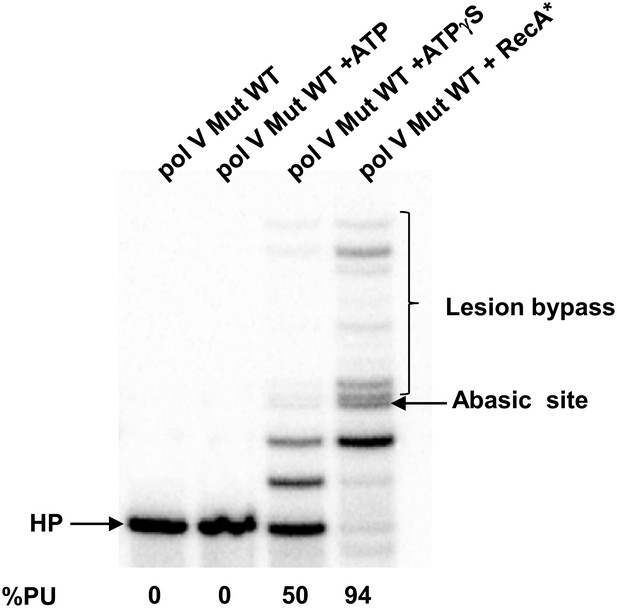
Pol V Mut WT activity on DNA containing an abasic site.
Pol V Mut (400 nM) activity was detected on 5′-32P-labeled 12 nt oh HP (100 nM), containing an abasic site 3 nts upstream from the 3′-OH, in the presence or absence of ATP/ATPγS and dNTPs. Lesion bypass is only observed when ATPγS is present.
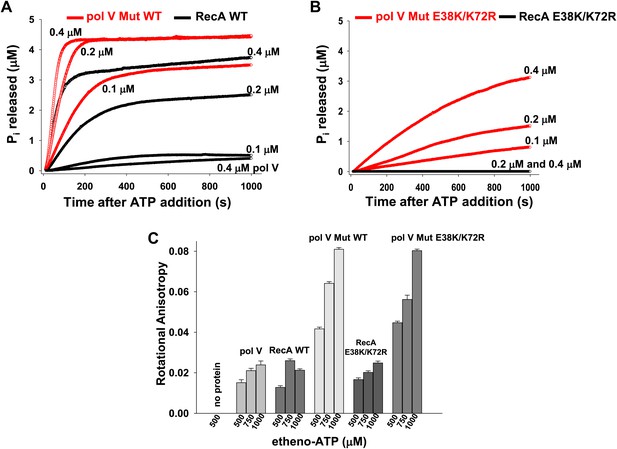
Pol V Mut is a DNA-dependent ATPase.
(A and B) ATP hydrolysis by pol V Mut and RecA (0.1 μM, 0.2 μM and 0.4 μM each) was measured using MDCC-PBP (5 μM) in the presence of 30 nt ssDNA (1 μM) and ATP (500 μM). MDCC-PBP fluorescence increases as Pi is released due to ATP hydrolysis. The measurements were taken at a resolution of 1 point per sec for approximately 1000 s. (C) Binding of 400 nM pol V, RecA and pol V Muts to etheno-ATP at varied concentrations was measured using rotational anisotropy. The error bars correspond to 1 SEM.
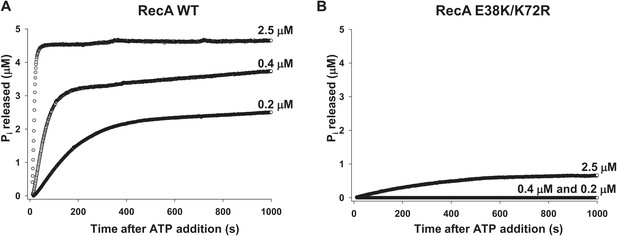
RecA WT and RecA E38K/K72R-dependent ATP hydrolysis.
ATP hydrolysis of (A) RecA WT and (B) RecA E38K/K72R was measured as a function of protein concentration in the presence of 1 µM 30 nt ssDNA and 500 µM ATP. Pi release resulting from ATP hydrolysis was measured as a change in fluorescence of MDCC-PBP (5 μM). The measurements were taken at 1 point per sec resolution for approximately 1000 s.
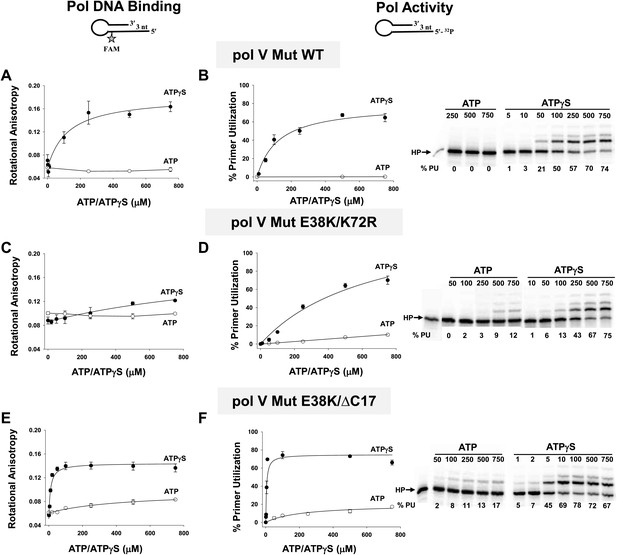
Pol V Mut binding and activity as a function of nucleotide.
Binding of pol V Mut WT (1 μM) (A), pol V Mut E38K/K72R (400 nM) (C), and pol V Mut E38K/ΔC17 (400 nM) (E) to 3 nt oh HP was measured as a change in rotational anisotropy. Activity of pol V Mut WT (400 nM) (B), pol V Mut E38K/K72R (400 nM) (D), and pol V Mut E38K/ΔC17 (400 nM) (F) was quantified on 5′-32P-labeled 3 nt oh HP with varying concentrations of nucleotide and 500 μM dNTPs. A gel showing primer utilization (%PU) as a function of nucleotide is presented to the right of the graph. ATP (open circle) and ATPγS (filled circle). The error bars correspond to 1 SEM.
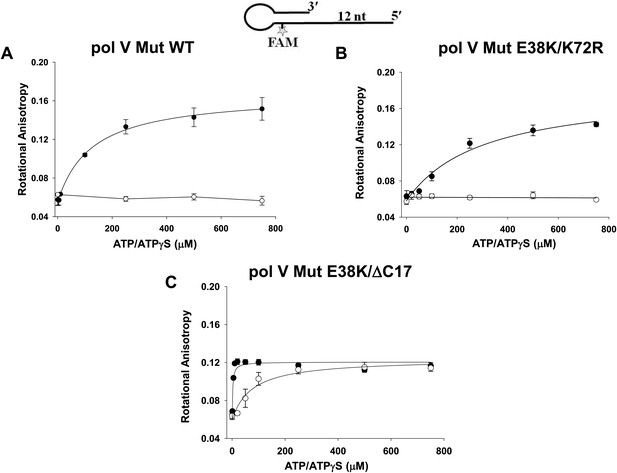
Pol V Mut binding to 12 nt oh HP DNA as a function of ATP/ATPγS.
Binding of (A) pol V Mut WT (B) pol V Mut E38K/K72R, and (C) pol V Mut E38K/ΔC17 (400 nM each) to fluorescein-labeled 12 nt oh HP (50 nM) was measured as a function of ATP (open circles) and ATPγS (filled circles). DNA binding was observed as a change in rotational anisotropy. The error bars correspond to 1 SEM.
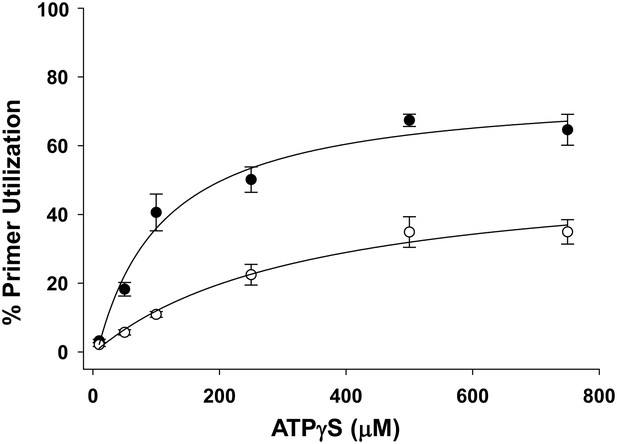
Pol V Mut WT activity as a function of ATPγS concentration in the presence of ATP.
Pol V Mut WT (400 nM) DNA synthesis is measured as a function of ATPγS (filled circles) on 3 nt oh HP (50 nM). When ATP (500 μM) is present at each ATPγS concentration, primer utilization of pol V Mut WT is decreased (open circles) compared to reactions where no ATP was included. The error bars correspond to 1 SEM.
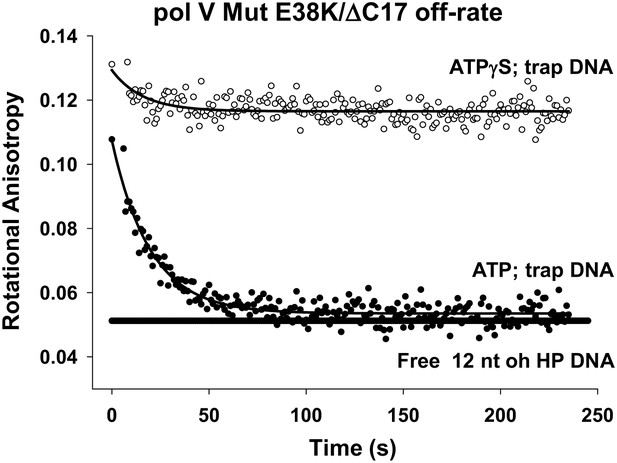
Pol V Mut E38K/ΔC17 off-rate in the presence of ATP and ATPγS.
Fluorescence depolarization of fluorescein-labeled (12 nt oh HP) DNA was used to measure the dissociation constant of pol V Mut E38K/ΔC17 in the presence of ATP (filled circles) and ATPγS (open circles). A stable protein–DNA complex (400 nM and 50 nM, respectively) was preformed in the presence of nucleotide followed by the addition of excess (160 times) trap DNA (unlabeled 12 nt oh HP). The decrease in anisotropy over time was fit to an exponential decay to determine koff (0.053 ± 0.0025 s−1).
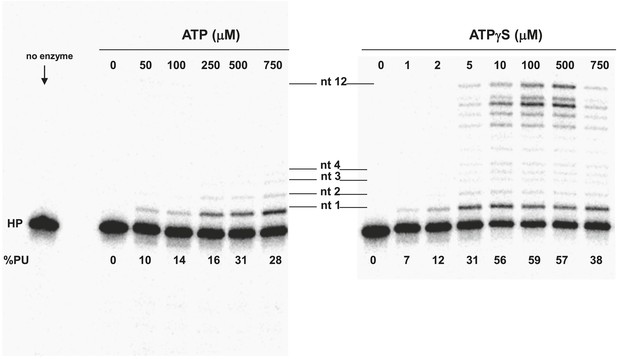
Pol V Mut E38K/ΔC17 primer extension length as a function of ATP or ATPγS concentration.
DNA extension was measured with varying concentrations of ATP (left panel) and ATPγS (right panel) on 12 nt oh HP to determine the lengths of primer synthesized as a function of NTPs. The unextended primer is indicated as HP and the addition of a nucleotide is marked as nt 1–4 and nt 12 in the center of the panels. The length of DNA synthesized by pol V Mut E38K/ΔC17 increases with the concentration of ATP/ATPγS. Pol V Mut E38K/ΔC17 is far more active with ATPγS compared with ATP, which is consistent with in inability to dissociate from p/t DNA in the absence of ATP hydrolysis.
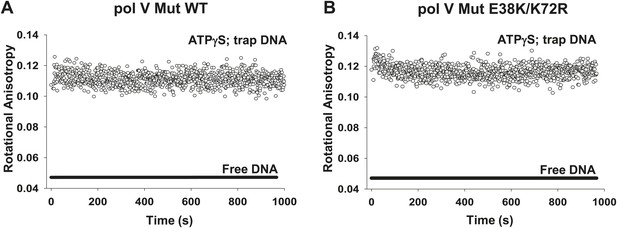
Dissociation of pol V Mut WT and pol V Mut E38K/K72R in the presence of ATPγS.
(A) pol V Mut WT and (B) pol V Mut E38K/K72R (400 nM) were pre-bound to fluorescein-labeled 12 nt oh HP (50 nM) in the presence of ATPγS. Excess trap DNA (8 μM unlabeled 12 nt oh HP) was added to the stable protein–DNA complex and DNA binding of pol V Mut was monitored as a change in rotational anisotropy (circles). Fluorescein-labeled 12 nt oh HP alone is indicated as a black line.
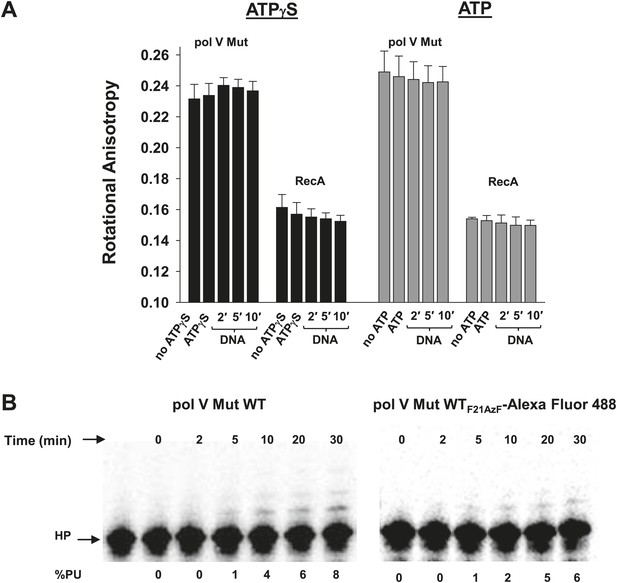
Pol V Mut remains intact in the presence of ATP/ATPγS and during DNA synthesis.
RecAF21AzF-Alexa Fluor 488 was used to form RecA* during pol V Mut assembly resulting in the formation of fluorescently-labeled pol V Mut (pol V Mut WTF21AzF-Alexa Fluor 488). (A) The rotational anisotropy of pol V Mut WTF21AzF-Alexa Fluor 488 (100 nM) was measured under ATP hydrolysis and DNA synthesis conditions (‘Materials and methods’). No change was observed upon addition of ATP/ATPγS, 12 nt oh HP (1 μM) or during DNA synthesis (2, 5 and 10 min) indicating that pol V Mut remains intact. The anisotropy of free RecAF21AzF-Alexa Fluor 488 (100 nM) was measured in parallel. The error bars correspond to 1 SEM. (B) DNA synthesis was measured for pol V Mut WT and pol V Mut WTF21AzF-Alexa Fluor 488 demonstrating that intact enzyme extends DNA under experimental conditions (pol V Mut: 100 nM and HP 1 μM) at the time periods (2, 5, 10, 20, and 30 min) used for rotational anisotropy measurements (A).
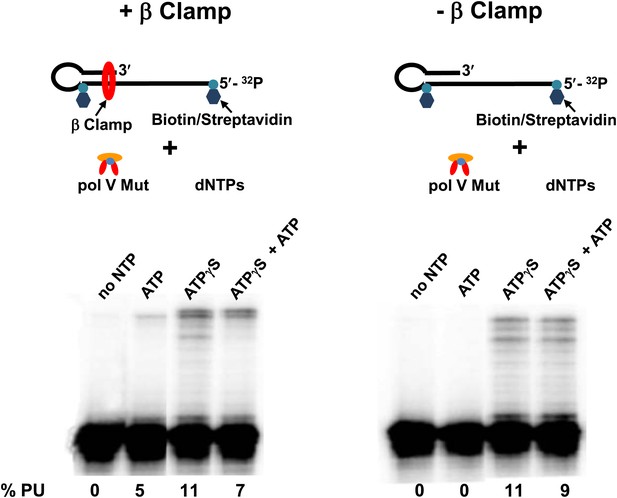
Pol V Mut WT is active with ATP only in the presence of β/γ complex.
Sketch of the experimental set up is illustrated above the gels. To prevent the β clamp from sliding off the DNA, a 12 nt oh HP was designed containing biotin/streptavidin on both sides of the primer terminus substrate. The activity of pol V Mut WT was measured in the presence and absence of β/γ. In the presence of β/γ complex (left panel) pol V Mut WT is able to extend p/t with ATP, in contrast no DNA synthesis is observed with ATP in the absence of β/γ complex (right panel).
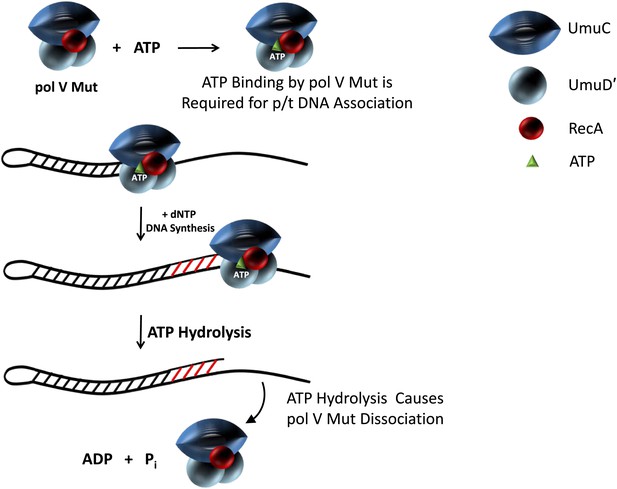
Model showing ATP regulation of pol V Mut activity.
Pol V Mut is active for DNA synthesis only after binding a molecule of ATP (green triangle) to form UmuD′2C-RecA-ATP. The binding of ATP is required for polymerase association with p/t DNA. ATP-hydrolysis catalyzed by an intrinsic DNA-dependent ATPase triggers pol V Mut-p/t DNA dissociation, while leaving intact the UmuD′2C-RecA complex.
Tables
Pol V Mut and RecA ATP hydrolysis rate constants
| ATPase kcat (s−1)* | |
|---|---|
| pol V Mut WT | |
| no DNA | (1.5 ± 0.1) × 10−3 |
| 12 nt oh HP | (9.0 ± 0.8) × 10−3 |
| 3 nt oh HP | (4.3 ± 0.1) × 10−3 |
| 30 nt ssDNA | (160 ± 5) × 10−3 |
| RecA WT | |
| 30 ssDNA | (100 ± 5) × 10−3 |
| pol V Mut E38K/K72R | |
| no DNA | (1.7 ± 0.2) × 10−3 |
| 12 nt oh HP | (4.4 ± 0.7) × 10−3 |
| 3 nt oh HP | (3.4 ± 0.7) × 10−3 |
| 30 nt ssDNA | (17 ± 1) × 10−3 |
| RecA E38K/K72R | |
| ssDNA | (0.6 ± 0.1) × 10−3† |
| pol V Mut E38K/ΔC17 | |
| no DNA | (7.0 ± 1.5) × 10−3 |
| 12 nt oh HP | (54 ± 9) × 10−3 |
| 3 nt oh HP | (46 ± 2) × 10−3 |
| 30 nt ssDNA | (90 ± 10) × 10−3 |
| RecA E38K/ΔC17 | |
| 30 nt ssDNA | (120 ± 15) × 10−3 |
-
*
kcat is an average of at least three independent measurements; ± SEM.
-
†
kcat was measured at 2.5 μM concentration for RecA E38K/K72R; ATP hydrolysis was not detectable at lower protein concentrations.
Pol V Mut affinity to 12 nt oh HP DNA
| Pol V Mut | Kd (nM) | |
|---|---|---|
| ATP | ATPγS | |
| pol V Mut WT | nb | 876 ± 52 |
| pol V Mut E38K/K72R | nb | 920 ± 112 |
| polV Mut E38K/ΔC17 | 312 ± 46 | 469 ± 27 |
-
nb–binding not detected.
Sequences for p/t HP DNA
| Sequences for p/t HP DNA | |
| 3 nt oh HP | 5′ AGA GCA GTT AGC GCA TTC AGC TCA TAC TGC TGA ATG CGC TAA CTG C 3′ |
| 3 nt oh HP (TTT) | 5′ TTT GCA GTT AGC GCA TTC AGC TCA TAC TGC TGA ATG CGC TAA CTG C 3′ |
| Fluorescein (FAM) 3 nt oh HP | 5′ AGA GCA GTT AGC GCA T(FAM)C AGC TCA TAC TGC TGA ATG CGC TAA CTG C 3′ |
| 12 nt oh HP | 5′ CGA AAC AGG AAA GCA GTT AGC GCA TTC AGC TCA TAC TGC TGA ATG CGC TAA CTG C 3′ |
| Fluorescein (FAM) 12 nt oh HP | 5′ CGA AAC AGG AAA GCA GTT AGC GCA TTC AGC TCA TAC TGC TGA A(FAM)G CGC TAA CTG C 3′ |
| Biotinylated (Bio) 12 nt oh HP | 5′ (Bio)GA AAC AGG AAA GCA GTT AGC GCA TTC AGC (Bio)CA TAC TGC TGA ATG CGC TAA CTG C 3′ |
| Sequences for pol V Mut assembly, activity and ATP hydrolysis DNA | |
| Amino C12-linked 45mer attached to cyanogen bromide-activated sepharose resin | 5′ C12TT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT T 3′ |
| ssDNA 30 mer for RecA transactivation and ATP hydrolysis | 5′ ACT GAC CCC GTT AAA ACT TAT TAC CAG TAA 3′ |





