Active invasion of bacteria into living fungal cells
Figures
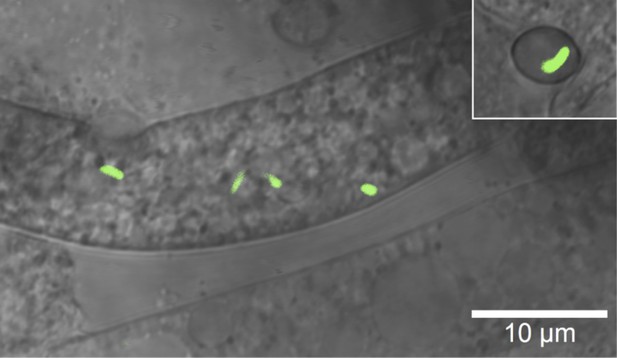
Microscopic image of Burkholderia rhizoxinica (green) residing in the cytosol of Rhizopus microsporus.
The GFP encoding B. rhizoxinica cells can re-colonize the sterile R. microsporus, then induce fungal sporulation. The endobacterium is transmitted via fungal vegetative spores (upper right corner).
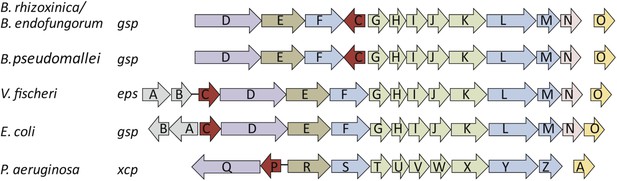
Schematic view of the organization of the type 2 secretion system (T2SS) gene clusters from various bacterial species.
The T2SS gene loci of the two R. microsporus endosymbiotic bacterial strains B. rhizoxinica and Burkholderia endofungorum, as well as the squid endosymbiont Vibrio fischeri, Escherichia coli, and the human pathogens Burkholderia pseudomallei and Pseudomonas aeruginosa are displayed here. The spaces between the arrows represent non-adjacent genes (single genes are located further away on the genome), while lines indicate closely linked genes.
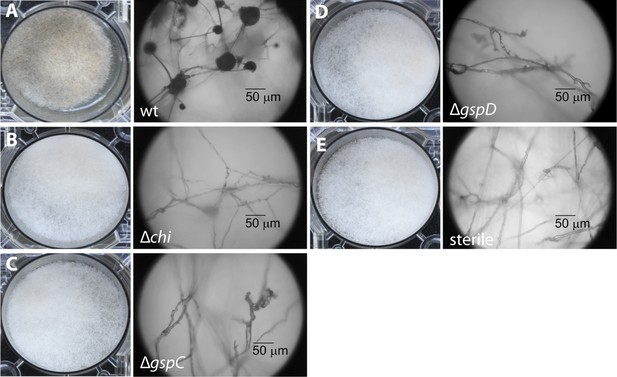
Photographs and microscopy images of R. microsporus hyphae several days after inoculation with B. rhizoxinica wt or mutant strains in 6-well plates.
The pictures present the infection of R. microsporus with wt or mutant B. rhizoxinica in the following order: (A) B. rhizoxinica wt, (B) B. rhizoxinica Δchi::Kanr, (C) B. rhizoxinica ΔgspC::Kanr, (D)—B. rhizoxinica ΔgspD::Kanr, (E)—control (no bacteria added). Spore formation is visible in (A), while spore formation was not detected in (B–E) even after 5 days of co-incubation.
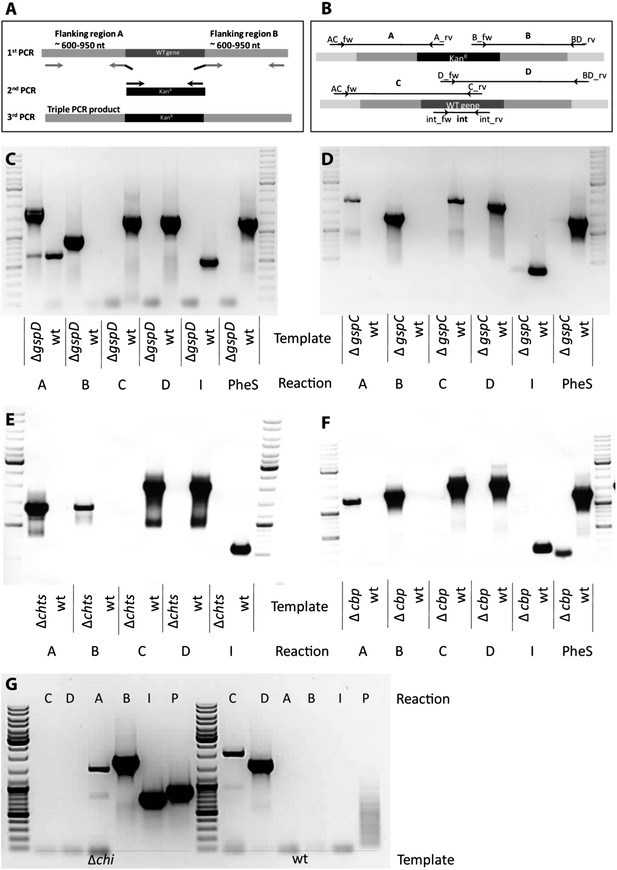
Knock out strategy of site directed mutagenesis to B. rhizoxinica.
(A)—Illustration of PCR strategy and primers used to confirm mutant genotypes. (B)—Confirmation of B. rhizoxinica ΔgspD::Kanr by gel electrophoresis of PCR products amplified from genomic DNA of B. rhizoxinica ΔgspD::Kanr or wt, respectively. Primers were combined in individual PCR reactions as indicated in above. (C) Confirmation of B. rhizoxinica ΔgspC::Kanr by PCR. (D) Confirmation of B. rhizoxinica Δchi::Kanr by PCR. (E) Confirmation of B. rhizoxinica Δcbp::Kanr by PCR. (F) Confirmation of B. rhizoxinica Δchts::Kanr by PCR. (G) Confirmation of B. rhizoxinica Δchi::Kanr by PCR.
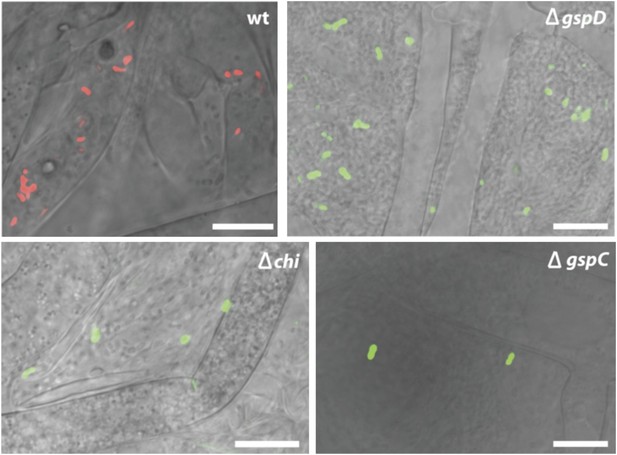
Fluorescence microscopy carried out with B. rhizoxinica wt and mutant strains B. rhizoxinica ΔgspD, B. rhizoxinica ΔgspC and B. rhizoxinica Δchi in a 3 day co-culture with sterile R. microsporus.
Whereas wt bacteria clearly localize within the fungal hyphae, no endobacteria carrying mutations in either the chitinase gene or the components of the T2SS are detectable within the living hyphae (upper part). Scale bar represents 10 µm.
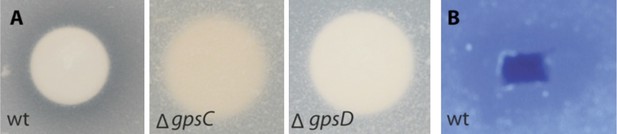
Lytic potential of B. rhizoxinica secretome.
(A)—Proteolytic assay using skim milk agar plates. Aliquots of the supernatants of B. rhizoxinica wt, B. rhizoxinica ΔgspD::Kanr, B. rhizoxinica ΔgspC::Kanr, respectively are incubated on filter disks on the plate. (B)—Chitinolytic plate assay using B. rhizoxinica culture supernatant on agar plate supplemented with 0.05% colloidal chitin and stained with Calcofluor.
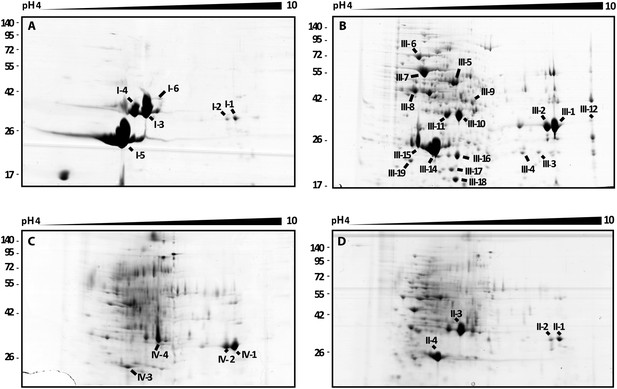
2D gel analysis of the secretomes of wild type and mutants.
(A) B. rhizoxinica wt, (B) B. rhizoxinica ΔgspC::Kanr, (C) B. rhizoxinica ΔgspD::Kanr and (D) B. rhizoxinica wt in co-culture with R. microsporus. Chitin-binding protein (I-5, III-14, III-15, IV-6, II-5), chitinase (I-6, III-9) and chitosanase (I-3, I-4, III-10, III-11, IV-3, IV-4, IV-5, II-3) were identified. List of identified proteins is shown in Table 2. In general, each gel was loaded with 100 μg of TCA-precipitated proteins from the culture supernatant.

Functional analyses of chitinolytic enzymes.
(A) Chitinase activity in cell-free culture supernatants of an E. coli harboring recombinant chitinase from B. rhizoxinica after incubation for 30, 60, and 90 min, in comparison to the activity of a recombinant S. lividans enzyme. Error bars indicate the standard deviation of three individual experiments. (B) Calcofluor staining of R. microsporus hyphae. (C) Phylogenetic analysis of chitinases from the GH family 18; protein sequences were retrieved from NCBI and comprise subfamily A and B sequences. (D) Chitin-binding assay performed using acid released crab shell chitin and the supernatant of B. rhizoxinica wt. SDS page of the non-bound fraction (S), the bound protein fraction (F1) and the pelleted chitin with the rest of the bound protein (F2). The three indicated proteins were identified using MALDI-TOF. (E) Gene expression assay for T2SS and chitinolytic proteins. The expression of T2SS genes gspC and gspD as well as chi, cbp and chts in B. rhizoxinica were monitored using RT qPCR in pure culture, in co-cultivation with a cured host (R. microsporus) and after re-infection of the cured host. The gene rpoB was used as an internal standard for the calculation of expression levels and normalization. The expression of all five genes is substantially increased in the wt during co-cultivation, while expression levels after re-infection decreased to nearly the level in pure wt culture. Error bars indicate standard deviation.
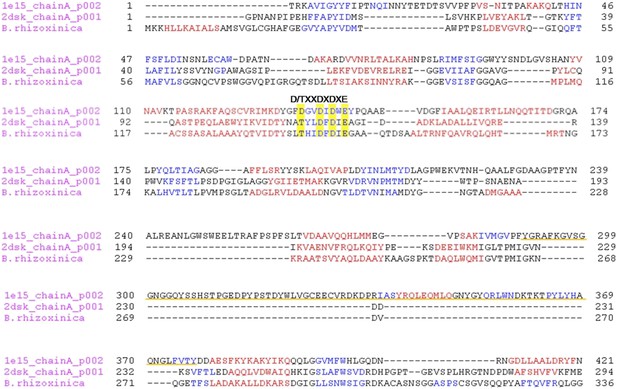
Multiple alignment of the chitinase protein sequences was performed based on the three-dimensional structure of the ChiB sequence from S. marcescens (pdb1e15) and Chi from P. furiosus (2dsk).
Red and blue characters indicate α-helices and β-strands, respectively. The DXDXE motif is highlighted in yellow. Alignment was created using PROMALS3D. The CID motif of subfamily A chitinases in S. marcescens in underlined in orange, it is absent in B. rhizoxinica and P. furiosus sequences.
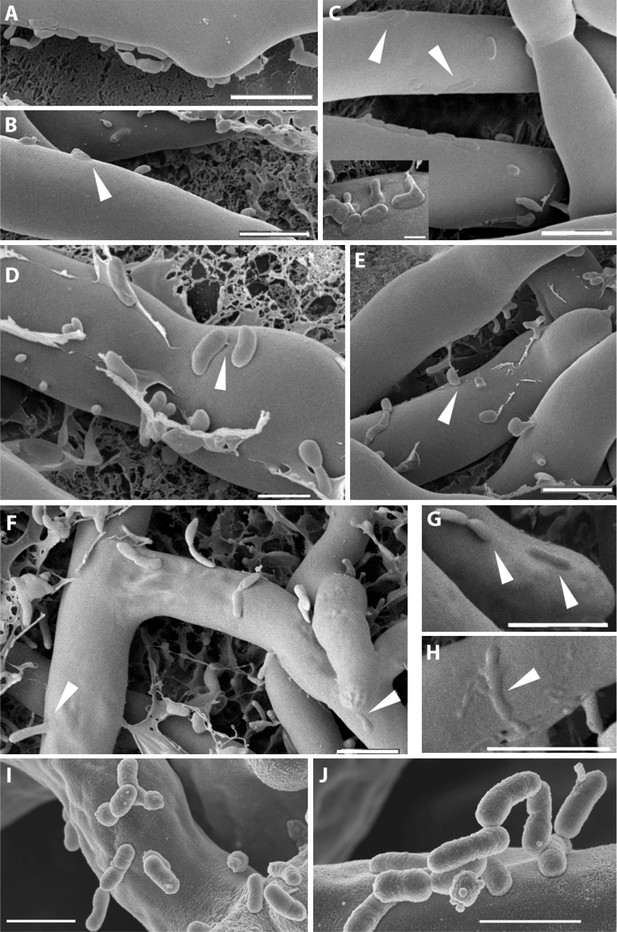
Course of infection of B. rhizoxinica (Br) to R. microsporus (Rm) observed by scanning cryo-electron microscopy.
(A–D) Attachment/adherence of bacteria to fungal hyphae after 1 hr and (F–H) 20 hr of co-cultivation; (C–D, F–H) bacterial and fungal cell walls start to merge; (D) fibrillar structures connecting a bacterial cell to the hyphal surface; (E–H) fusion of cell walls and the intrusion of bacterial cells into the fungal hyphae. White arrows mark areas of particular interest. (I–J) Scanning electron microscopy of a co-culture of sterile R. microsporus with B. rhizoxinica ΔgspD (I) and B. rhizoxinica Δchi (J). Attachment of both mutant strains to the hyphal surface is visible, however, no intimate contact or fusion events could be observed. Scale bars represent 5 µm.
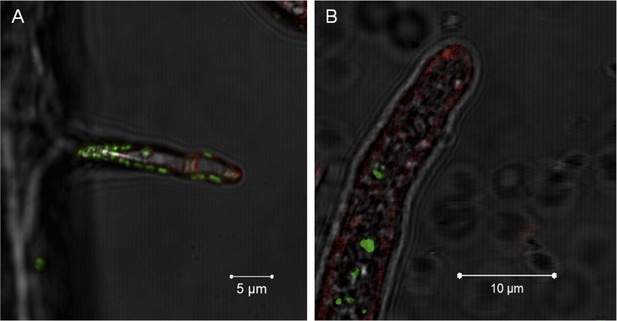
Evidence for the lack of active engulfment of B. rhizoxinica by R. microsporus.
(A) Bacteria (green) attaches to fungal hyphae within 1 hr. (B) The endocytotic activity of the fungus can be observed by the red vesicles that are present all around the hyphae and highly accumulated at the apical tip. After the infection, no fungal membrane is visible around the bacterium.
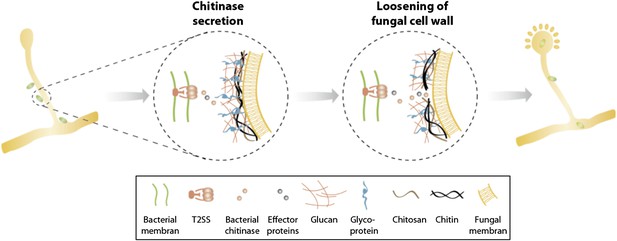
Model of processes involved in bacterial invasion.
Chitinase as well as other effector proteins are secreted via a bacterial T2SS and induce a local dissolution of the fungal cell wall. This enables bacteria to enter and colonize the fungal cell and induce sporulation.
Tables
Annotation of the B. rhizoxinica T2SS gene cluster
| Gene | Proposed function of encoded protein | Homolog in Klebsiella oxytoca | Percent identity (BlastP) | Homolog in Burkholderia pseudomallei | Percent identity (BlastP) |
|---|---|---|---|---|---|
| gspC | Connecting inner and outer membrane complex | pulC | – | gspC | 61 |
| gspD | Secretin, outer membrane pore formation | pulD | 39 | gspD | 64 |
| gspE | Cytoplasmitc ATPase, energy for translocation of pseudopilins | pulE | 54 | gspE | 89 |
| gspF | Anchoring protein, inner membrane platform for pseudopilins | pulF | 46 | gspF | 83 |
| gspG | Major prepilin-like protein, pilus-like structure formation | pulG | 56 | gspG | 87 |
| gspH | Pseudopilin subunit, pilus-like structure formation | pulH | 70 | gspH | 66 |
| gspI | Pseudopilin subunit, pilus-like structure formation | pulI | 44 | gspI | 66 |
| gspJ | Pseudopilin subunit, pilus-like structure formation | pulJ | 43 | gspJ | 55 |
| gspK | Pseudopilin subunit, pilus-like structure formation | pulK | 26 | gspK | 62 |
| gspL | Anchoring protein, inner membrane platform for pseudopilins | pulL | 27 | gspL | 57 |
| gspM | Anchoring protein, inner membrane platform for pseudopilins | pulM | 24 | gspM | 56 |
| gspN | Connecting inner and outer membrane complex | pulN | 29 | gspN | 63 |
| gspO | Prepilin, inner membrane peptidase | pulO | 43 | gspO | 60 |
Secretome proteins identified by MALDI-TOF
| Spot no. | Accesion no. (NCBI) | Protein indetification | Total Mass0 (kDA) | Total pI | Mascot score | Matching peptides | Sequence coverage (%) | SignalP prediction (y/n) |
|---|---|---|---|---|---|---|---|---|
| I-1 | gi│312169059 | Glutamate/aspartate-binding protein | 32.4 | 9.6 | 64.2 | 13 | 40.1 | y |
| I-2 | gi│312169059 | Glutamate/aspartate-binding protein | 32.4 | 9.6 | 114.0 | 18 | 49.5 | y |
| I-3 | gi│312167773 | Chitosanase (EC 3.2.1.132) | 39.6 | 6.5 | 88.3 | 10 | 38.9 | y |
| I-4 | gi│312167773 | Chitosanase (EC 3.2.1.132) | 39.6 | 6.5 | 124.0 | 15 | 47.7 | y |
| I-5 | gi│312168534 | Chitin-binding protein | 27.0 | 7.7 | 85.2 | 8 | 49.2 | y |
| I-6 | gi│312168091 | Chitinase (EC 3.2.1.14) | 42.1 | 8.7 | 30.8 | 6 | 23.1 | y |
| II-1 | gi│312169059 | Glutamate/aspartate-binding protein | 32.4 | 9.6 | 107.0 | 20 | 51.5 | y |
| II-2 | gi│312169059 | Glutamate/aspartate-binding protein | 32.4 | 9.6 | 114.0 | 18 | 46.8 | y |
| II-3 | gi│312167773 | Chitosanase (EC 3.2.1.132) | 39.6 | 6.5 | 169.0 | 15 | 60.3 | y |
| II-4 | gi│312168534 | Chitin-binding protein | 27.0 | 7.7 | 80.4 | 7 | 29.1 | y |
| III-1 | gi│312169059 | Glutamate/aspartate-binding protein | 32.4 | 9.6 | 133.0 | 20 | 51.9 | y |
| III-2 | gi│312169059 | Glutamate/aspartate-binding protein | 32.4 | 9.6 | 96.2 | 19 | 51.2 | y |
| III-3 | gi│312167620 | Toluene transport system Ttg2d protein | 23.7 | 9.5 | 69.0 | 5 | 30.5 | y |
| III-4 | gi│312168022 | Ribosome recycling factor (RRF) | 21.0 | 9.0 | 74.0 | 9 | 44.1 | n |
| III-5 | gi│312169310 | Adenosylhomocysteinase (EC 3.3.1.1) | 52.7 | 5.9 | 247.0 | 29 | 69.2 | n |
| III-6 | gi│312166966 | Chaperone protein DnaK | 69.7 | 4.9 | 222.0 | 30 | 48.2 | n |
| III-7 | gi│312168888 | 60 kDa chaperonin GroEL | 57.4 | 5.1 | 97.7 | 16 | 41.0 | n |
| III-8 | gi│312169323 | S-adenosylmethionine synthetase (EC 2.5.1.6) | 42.8 | 4.8 | 182.0 | 20 | 63.8 | n |
| III-9 | gi│312167529 | Protein translation elongation factor Tu (EF-Tu) | 43.1 | 5.2 | 148.0 | 22 | 66.9 | n |
| III-10 | gi│312167773 | Chitosanase (EC 3.2.1.132) | 39.6 | 6.5 | 156.0 | 17 | 63.6 | y |
| III-11 | gi│312167773 | Chitosanase (EC 3.2.1.132) | 39.6 | 6.5 | 127.0 | 16 | 54.0 | y |
| III-12 | gi│312168091 | Chitinase (EC 3.2.1.14) | 42.1 | 8.7 | 42.8 | 8 | 29.1 | y |
| III-13 | gi│312168185 | Peptidyl-prolyl cis-trans isomerase (EC 5.2.1.8) | 28.7 | 8.9 | 91.0 | 12 | 40.8 | y |
| III-14 | gi│312168534 | Chitin-binding protein | 27.0 | 7.7 | 101.0 | 12 | 59.8 | y |
| III-15 | gi│312168534 | Chitin-binding protein | 27.0 | 7.7 | 60.6 | 5 | 34.8 | y |
| III-16 | gi│312167051 | Superoxide dismutase (EC 1.15.1.1) | 23.5 | 5.9 | 128.0 | 9 | 58.7 | n |
| III-17 | gi│312168185 | Peptidyl-prolyl cis-trans isomerase (EC 5.2.1.8) | 28.7 | 8.9 | 84.4 | 13 | 37.6 | y |
| III-18 | gi│312167149 | Inorganic pyrophosphatase (EC 3.6.1.1) | 19.4 | 4.8 | 117.0 | 12 | 84.1 | n |
| III-19 | gi│312167795 | 34 kDa membrane antigen precursor | 21.8 | 6.8 | 85.5 | 13 | 60.7 | y |
| IV-1 | gi│312169059 | Glutamate/aspartate-binding protein | 32.4 | 9.6 | 151.0 | 23 | 62.6 | y |
| IV-2 | gi│312169059 | Glutamate/aspartate-binding protein | 32.4 | 9.6 | 98.6 | 15 | 45.5 | y |
| IV-3 | gi│312167773 | Chitosanase (EC 3.2.1.132) | 39.6 | 6.5 | 71.3 | 8 | 23.0 | y |
| IV-4 | gi│312168534 | Chitin-binding protein | 27.0 | 7.7 | 95.3 | 7 | 37.3 | y |
Bacterial and fungal strains
| Stains | Characteristics | References |
|---|---|---|
| Burkholderia rhizoxinica HKI-0454 | Wild type, isolated from Rhizopus microsporus ATCC62417 | * |
| Rhizopus microsporus ATCC62417 | Fungal host harboring bacterial endosymbionts, isolated from rice seedlings | † |
| B. rhizoxinica ΔgspD::Kanr | T2SS mutant B. rhizoxinica with deletion of gspD | This study |
| B. rhizoxinica ΔgspC::Kanr | T2SS mutant B. rhizoxinica with deletion of gspC | This study |
| B. rhizoxinica Δchit::Kanr | B. rhizoxinica with deletion of chitinase gene | This study |
| B. rhizoxinica Δcbp::Kanr | B. rhizoxinica with deletion of chitin-binding protein gene | This study |
| B. rhizoxinica Δchts::Kanr | B. rhizoxinica with deletion of chitosanase gene | This study |
| B. rhizoxinica/pHKT4 | B. rhizoxinica wt harboring a RFP expression vector | This study |
| B. rhizoxinica ΔgspD::Kanr/pHKT2 | B. rhizoxinica gspD mutant harboring a GFP expression vector | This study |
| B. rhizoxinica ΔgspC::Kanr/pHKT2 | B. rhizoxinica gspC mutant harboring a GFP expression vector | This study |
| B. rhizoxinica Δchit::Kanr/pHKT2 | B. rhizoxinica chit mutant harboring a GFP expression vector | This study |
| B. rhizoxinica Δcbp::Kanr/pHKT2 | B. rhizoxinica cbp mutant harboring a GFP expression vector | This study |
| B. rhizoxinica Δchts::Kanr/pHKT2 | B. rhizoxinica chts mutant harboring a GFP expression vector | This study |
| R. microsporus + B. rhizoxinica ΔgspD::Kanr/pHKT2 | Reinfected cured fungal host with gspD mutant harboring a GFP expressing vector | This study |
| R. microsporus + B. rhizoxinica ΔgspC::Kanr/pHKT2 | Reinfected cured fungal host with gspC mutant harboring a GFP expressing vector | This study |
| R. microsporus + B. rhizoxinica Δchit::Kanr/pHKT2 | Reinfected cured fungal host with chit mutant harboring a GFP expressing vector | This study |
| R. microsporus + B. rhizoxinica Δcbp::Kanr/pHKT2 | Reinfected cured fungal host with cbp mutant harboring a GFP expressing vector | This study |
| R. microsporus + B. rhizoxinica Δchts::Kanr/pHKT2 | Reinfected cured fungal host with chts mutant harboring a GFP expressing vector | This study |
| Escherichia coli BL21(DE3)/pET28a-Chi | E. coli with expression vector harboring chitinase gene | This study |
-
*
Partida-Martinez and Hertweck (2005) Pathogenic fungus harbours endosymbiotic bacteria for toxin production. Nature 437:884–888.
-
†
Ibaragi (1973) Studies on rice seedling blight. I. Growth injury caused by Rhizopus sp. under high temperature. Ann. Phytopathol. Soc. Jpn 39:141–144.
Primers used in this study
| Name | Oligo sequence | |
|---|---|---|
| Primers used for generating KO mutants | TII_D_fl1_fw | 5′-GCTACGGATCCCTGCCAGGTATTGCCGTATT-3′ |
| TII_D_fl1_rv | 5′-GCTACAAGCTTCAATCAGCTTGTCGAATTGC-3′ | |
| TII_D_fl2_rv | 5′-GCCCAGTAGCTGACATTCATCCCCGATCAATTATGCAAGCAG-3′ | |
| TII_D_fl2_fw | 5′-TTCTTGACGAGTTCTTCTGATGGACTGGATGTCTGGATCA-3′ | |
| TII_C_fl1_fw | 5′-GCTACGAATCTCAGATCTGTGCGAGGATTG-3′ | |
| TII_C_fl1_rv | 5′-GCTACAAGCTTCAACTCGCCTTTACGTACCC-3′ | |
| TII_C_fl2_rv2 | 5′-GCCCAGTAGCTGACATTCATCCGACGGCATGATGAGTTTGTG-3′ | |
| TII_C_fl2_fw2 | 5′-TTCTTGACGAGTTCTTCTGAAGCAAGCTGGTCAGGAACAT-3′ | |
| Kan_F | 5′-ATGATTGAACAAGATGGATTGC-3′ | |
| Kan_R | 5′-GCCTTCTTGACGAGTTCTTCTGA-3′ | |
| Chi_Fl1_F | 5′-GAACTAGTCTCGATCATGGGGGTATTTG-3′ | |
| Chi_Fl1_R | 5′-GCCCAGTAGCTGACATTCATCCCAGGTGCTTTTTCATTGCTTC-3′ | |
| Chi_Fl2_F | 5′-GCCTTCTTGACGAGTTCTTCTGACGTGACGTATCGTGCAAAGT-3′ | |
| Chi_Fl2_R | 5′-ATCCCGGGACGCGGTCAAGTCGATGTAG-3′ | |
| Kan_Chi_F | 5′-GAAGCAATGAAAAAGCACCTGGGATGAATGTCAGCTACTGGGC-3′ | |
| Kan_Chi_R | 5′-ACTTTGCACGATACGTCACGTCAGAAGAACTCGTCAAGAAGGC-3′ | |
| P1_chtos | 5′-GCTACGGGCCCGGCATCGGTGACTATCGTAAC-3′ | |
| P2_chtos | 5′-GCTACTTAATTAAGCTAGCGTAGCACAGCCGATACCGTAAGC-3′ | |
| P3_chtos | 5′-GCTACGCTAGCTTAATTAAGTAGCGCAATGGAGCAAGCTGATGG -3′ | |
| P4_chtos | 5′-GCTACGCGGCCGCAACGTGCGCGACGATACGTTC-3′ | |
| Kan_chts_F | 5′-GGATGAATGTCAGCTACTGGGC-3′ | |
| Kan_chts_R | 5′-TCAGAAGAACTCGTCAAGAAGGC-3′ | |
| P1_chtbdp | 5′-GCTACGCGGCCACGCCGAGATGATGTTG-3′ | |
| P2_chtbdp | 5′-GCTACTTAATTAAGCTAGCGTAGCCGATCGTGCGTGAGTAAG-3′ | |
| P3_chtbdp | 5′-GCTACGCTAGCTTAATTAAGTAGCAGCCAACCGACGTACCTACC-3′ | |
| P4_chtbdp | 5′-GCTACGGGCCCAAGACGGCGGGCGTATTACC-3′ | |
| Kan_cbp_F | 5′-GGATGAATGTCAGCTACTGGGC-3′ | |
| Kan_cbp_R | 5′-TCAGAAGAACTCGTCAAGAAGGC-3′ | |
| Primers used for RT-qPCR studies | TIISS_D_RT_F | 5′-GAGCAGCGATACCAACATCC-3′ |
| TIISS_D_RT_R | 5′-TTGAATGCGGAGACCGAAG-3′ | |
| TIISS_C_RT_F | 5′-AGCGTCACTTACTGGGTCATC-3′ | |
| TIISS_C_RT_R | 5′-CGAGCCGAACAGAGTTTGAG-3′ | |
| Chi_RT_F | 5′-CGCTGGATACGGTCAACATC-3′ | |
| Chi_RT_R | 5′-GCCTTGCACGTCATTCTT-3′ | |
| CBP_RT_F | 5′-ACGACAGCGCATAATCCTTC-3′ | |
| CBP_RT_R | 5′-GGGTGCATCGTAAATCAGGT | |
| Chtos_RT_F | 5′-AGGTGGACTGACCCGTATTG | |
| Chtos_RT_R | 5′-TTGCACGCTGTATTGGATGT-3′ | |
| rpoB_RT_F | 5′-ATTTCCTTCACCAGCACGTT-3′ | |
| rpoB_RT_R | 5′-TTCGGGGAAATGGAAGTGT-3′ | |
| Primers used for control of generated mutants | Arm_A_rv (Kan) | 5′-AGTGACAACGTCGAGCACAG-3′ |
| Arm_B_fw (Kan) | 5′-CGTTGGCTACCCGTGATATT-3′ | |
| TIISSD_C_A_fw | 5′-TCACCTCACGTAGCAGATCG-3′ | |
| TIISSD_C_rv | 5′-GCATCGACGAAATTCAAGGT-3′ | |
| TIISSD_D_fw | 5′-GATAACCGGATCGTCAAGGA-3′ | |
| TIISSD_D_B_rv | 5′-CCGGACAAGTCGTACTCGAT-3′ | |
| TIISSD_Int_fw | 5′-GTCGAGGGACCAAAGTTTCA-3′ | |
| TIISSD_Int_rv | 5′-GGCGTAGACAGGATGTTGGT-3′ | |
| TIISSC_CA_fw | 5′-ACTCCAGCCCGCATACATAC-3′ | |
| TIISSC_C_rv | 5′-ATTCAGCGCACGTAGATCGT-3′ | |
| TIISSC_D_fw | 5′-GCGTCACTTACTGGGTCATC-3′ | |
| TIISSC_DB_rv | 5′-AGGAAGTGCTGCGTGTAACC-3′ | |
| Chi_Ctrl1_KO_F | 5′-GAACCATTCGCCTTCTTCAC-3′ | |
| Chi_Ctrl1_WT_R | 5′-ATCGCTTTCAACAGGTGCTT-3′ | |
| Chi_Ctrl2_WT_R | 5′-CCAGTTGTGGCAAATGATTG-3′ | |
| Chi_Ctrl2_KO_R | 5′-ATTTCGGCTCTGACGTGACT-3′ | |
| Chi_Int_fw | 5′-TGACCTCCATCGCCAAGTCG-3′ | |
| Chi_Int_rv | 5′-CGGAACACCTGCGTGAATGC-3′ | |
| Chtos_ext_for_1 | 5′-GAAGCGTGATGTGATTGAAG-3′ | |
| Chtos_ext_rev_1 | 5′-AAGTCGCATCCAGACATTG-3′ | |
| Chtos_int_for_1 | 5′-GACGCCAAGACGATCTACCA-3′ | |
| Chtos_int_rev_1 | 5′-TTGGGCTTTGACCTTGCTAC-3′ | |
| Cbp_ext_for_1 | 5′-ACTTTCTGAATACAGCTTGC-3′ | |
| Cbp_ext_rev_1 | 5′-CAGTCATGATGCAATACGTG-3′ | |
| Cbp_int_for_1 | 5′-GCGGTCTAGTCCCTGCTTAC-3′ | |
| Cbp_int_rev_1 | 5′-GAGGCTATTGGTCGTCACCT-3′ | |
| pBS_nspI_for_I | 5′-AGCTCACTCAAAGGCGGTAA-3′ | |
| pBS_nspI_rev_I | 5′-TTTTTGTGATGCTCGTCAGG-3′ |





