Noncanonical binding of BiP ATPase domain to Ire1 and Perk is dissociated by unfolded protein CH1 to initiate ER stress signaling
Figures
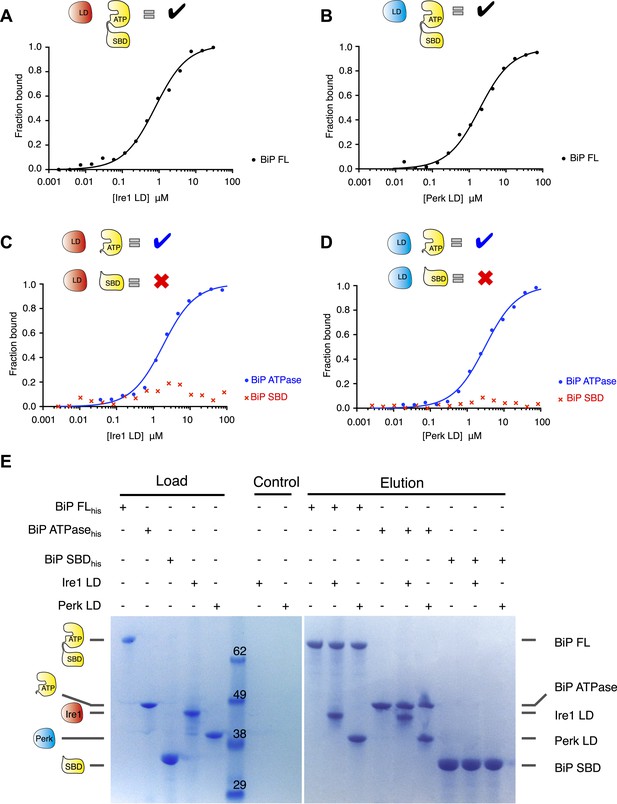
Noncanonical binding of BiP ATPase domain to Ire1 and Perk.
(A–B) Microscale thermophoresis (MST) analysis showing sigmoidal binding curves for interaction between full-length BiP and the complete luminal domains (region I–V) of (A) Ire1 luminal domain (Kd = 1.33 μM) and (B) Perk luminal domain (Kd = 1.92 μM). (C–D) MST binding curves of interaction between BiP sub-domains (ATPase and substrate binding domain) and (C) Ire1 luminal domain (ATPase Kd = 1.97 μM; no binding to substrate binding domain) and (D) Perk luminal domain (ATPase Kd = 2.05 μM; no binding to substrate binding domain). (E) Pull down assay showing BiP-luminal domain complexes using His6-tagged BiP proteins and luminal domains of Perk and Ire1 visualized by coomassie brilliant blue stained SDS PAGE gel. Ire1 and Perk luminal domains bind to full-length BiP and BiP ATPase domain. No binding to BiP substrate binding domain was observed.
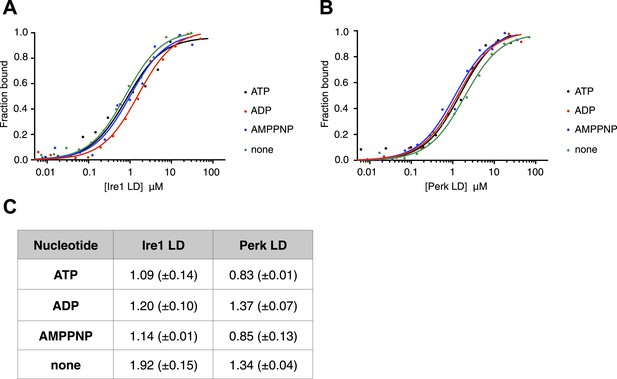
The noncanonical binding of BiP ATPase domain to Ire1 and Perk is independent of nucleotides.
(A–B) MST analysis showing sigmoidal binding curves for full-length BiP interaction with (A) Ire1 luminal domain and (B) Perk luminal domain in the presence of 10 mM ATP, ADP, AMPPNP and in the absence of nucleotides. (C) List of Kd values (μM ± SE) for Ire1 and Perk luminal domain interactions with full-length BiP in the presence of nucleotides for the binding curves represented in A and B. Binding between luminal domains and BiP was not affected by the presence of the various nucleotides.
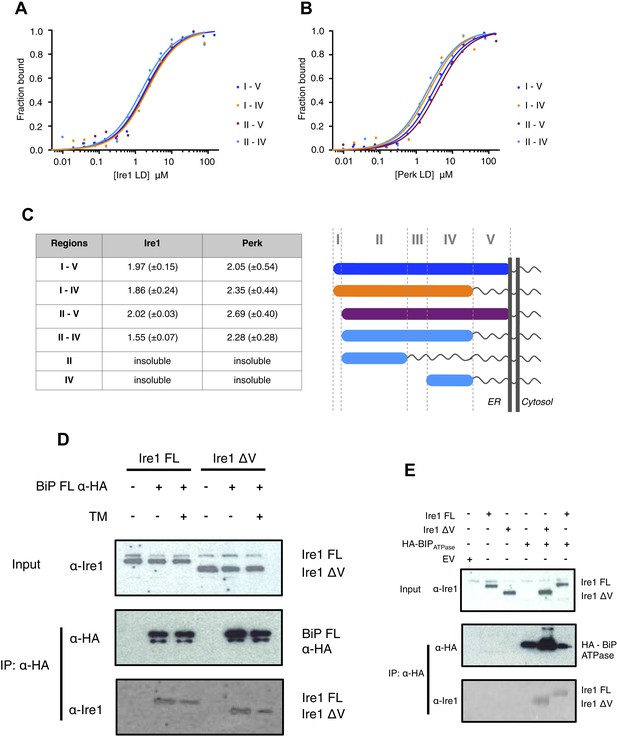
Core interaction between BiP ATPase and luminal domains occurs via region II-IV of luminal domains.
(A–B) MST binding curves of interaction between BiP ATPase and different length constructs of (A) Ire1 luminal domain and (B) Perk luminal domain. (C) List of Kd values (μM ± SE) for BiP ATPase interaction with the various Ire1 and Perk luminal domain constructs (based on regions I–V) for binding curves represented in A and B. The luminal domain region II–IV, is solely responsible for binding to BiP proteins and regions I and V are dispensable in this interaction. (D) Co-immunoprecipitation experiment in which HEK293T cells were co transfected with either Ire1 mutant lacking region V (Ire1ΔV; Δ390–430) or full-length Ire1, along with HA-tagged BiP, in the absence or presence of ER stress (TM = 5 μM; 4 hr tunicamycin). Immunoprecipitating with HA peptide and then immunoblotting with Ire1 specific antibody reveals an interaction between BiP and both full-length and mutant Ire1 that is missing region V (Ire1 ΔV), which is reduced after ER stress. This interaction in cells reinforces the in vitro data. (E) Co-immunoprecipitation experiment similar to (D), but cells were co transfected with HA-tagged BiP ATPase and were not subjected to ER stress with tunicamycin.
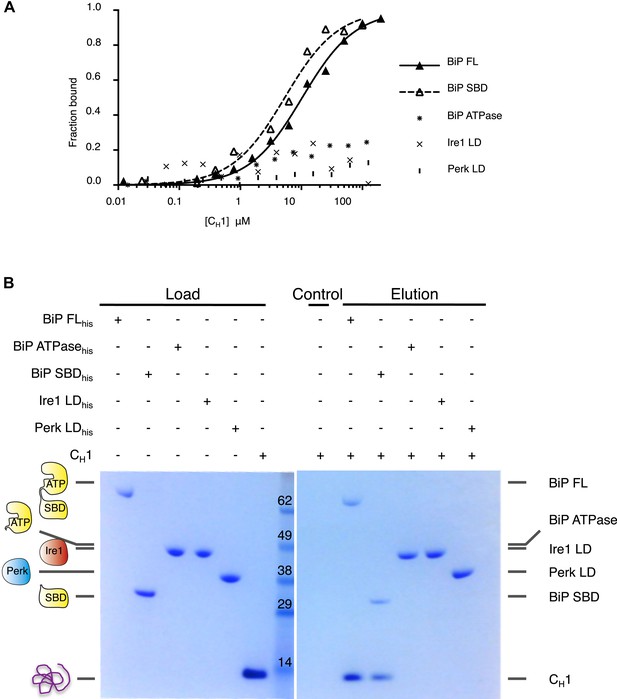
Unfolded protein CH1 binds to canonical BiP substrate binding domain without binding to UPR luminal domains.
(A) MST binding curves for CH1 binding to full-length BiP (Kd = 8.7 μM), BiP's ATPase domain (no binding), BiP's substrate binding domain (Kd = 5.1 μM), Ire1 luminal domain (no binding) and Perk luminal domain (no binding). (B) Pull down experiment showing CH1 binding to both full-length and substrate binding domain of BiP only, with no interaction observed to luminal domains, reaffirming the data for CH1 interactions using MST in part A.
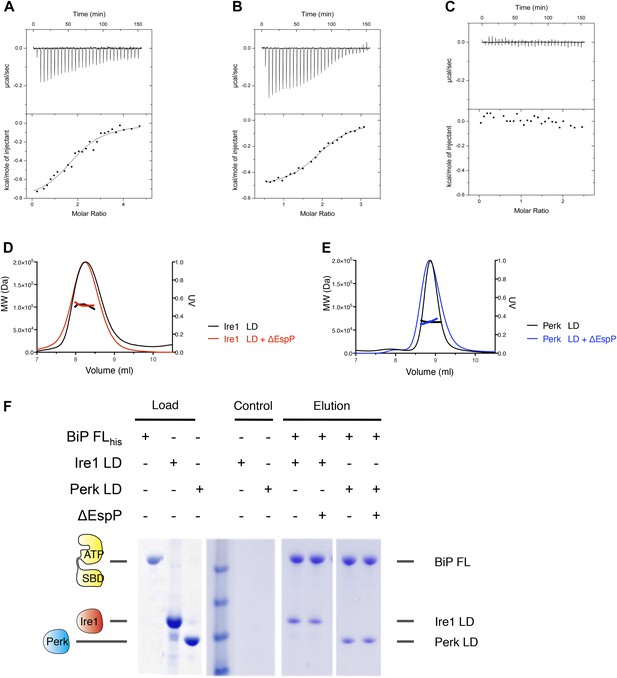
Assessing the role of unfolded protein peptide mimic (ΔEspP) in UPR stress sensing.
(A–C) ITC binding curves for ΔEspP titration into (A) Ire1 luminal domain (Kd = 6.4 μM; N = 1.97), (B) Perk luminal domain (Kd = 9.3 μM; N = 1.98), (C) full-length BiP (no binding). (D) SEC MALS analyses showing Ire1 luminal domain is dimeric in solution (MW = 104.5 kDa) and the addition of ΔEspP has no effect on Ire1 luminal domain oligomeric state (MW = 103.2 kDa). (E) SEC MALS analyses showing Perk luminal domain is dimeric in solution (MW = 67.8 kDa) and the addition of ΔEspP has no effect on Perk luminal domain oligomeric state (MW = 67.6 kDa). (F) Pull down assay to test the effect of ΔEspP on His6-tagged full-length BiP-luminal domain complexes. ΔEspP has no visible effect upon BiP-luminal domain complexes.
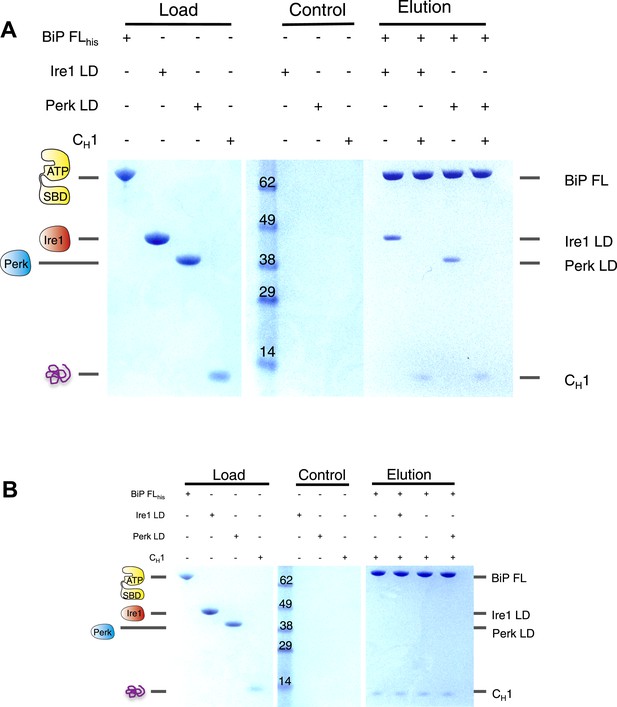
The unfolded protein CH1 dissociates the noncanonical interaction between BiP ATPase domain and the luminal domain of Ire1 or Perk.
(A) Pull down assay assessing the effects upon addition of unfolded protein CH1 to His6-tagged full-length BiP-luminal domain complexes. CH1 disrupts BiP-luminal domain interaction and causes the complexes to dissociate. (B) When His6-tagged full-length BiP is initially incubated with CH1 and then subsequently Ire1 and Perk luminal domains are added, we see no binding between luminal domains and BiP indicating that luminal domains and CH1 binding to BiP are mutually exclusive.
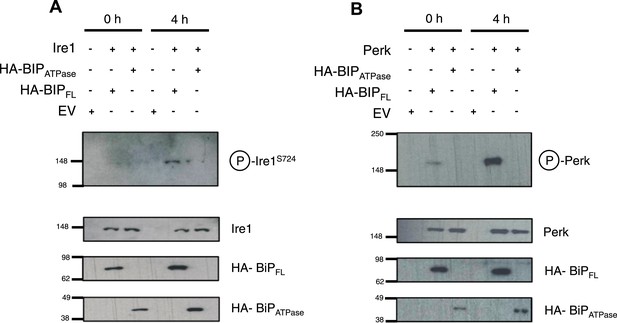
BiP deletion mutants, lacking the substrate binding domain, attenuate UPR signaling.
(A) Ire1 was co expressed with either full-length BiP (HA–BiPFL) or BiP ATPase domain, lacking the substrate binding domain (HA–BiPATPase), and challenged with tunicamycin (5 μM) over 0 hr and 4 hr time points in Ire1−/− cells. S724 phosphorylated Ire1 was measured as an indicator of UPR signaling (Ali et al., 2011) using pIre1s724 antibody (ICR). Cells expressing full-length BiP and Ire1 were fully able to respond to induced ER stress, whilst expression with BiP ATPase domain attenuated UPR signaling. EV = empty vector. (B) Similar to (A), but expressing Perk full-length in Perk−/− cells and using pPerk antibody (Santa Cruz).
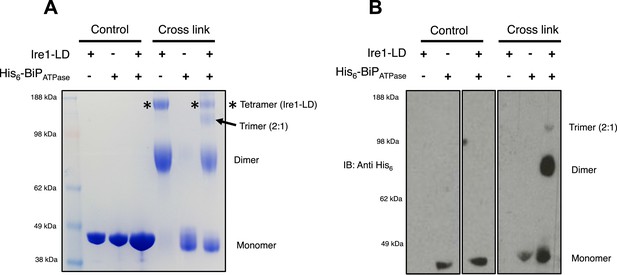
BiP impedes Ire1 LD dimer and tetramer formation.
(A) Ire1 luminal domain (LD; regions II–IV) and His6-tagged BiP ATPase domain proteins, both individually and in 1:1 molar ratio mixture, were visualized as control lanes on a 4–12% Bis-Tris SDS-PAGE gel. The same proteins were then subjected to EGS cross linker for 1 hr, after which the reaction was quenched and samples were visualized along side control lanes. In the presence of cross linker, Ire1 LD forms dimer and tetramer species; when in a mixture with BiP ATPase, there is a reduction in the corresponding tetramer band (*). Also, a band appears that is consistent in size with a trimer species. (B) Samples from (A) were immunoblotted using anti-His6 antibody, which detects His6-tagged BiP ATPase domain protein. Since BiP ATPase protein is monomeric in the absence or presence of cross linker, BiP ATPase forms hetero dimer and hetero trimer with Ire1 LD. The binding of BiP to Ire1 reduces the size of the tetramer (*) that is exclusively formed by Ire1 LD, leading to the conclusion that BiP inhibits Ire1 LD tetramer formation, by preventing formation of the dimer species.
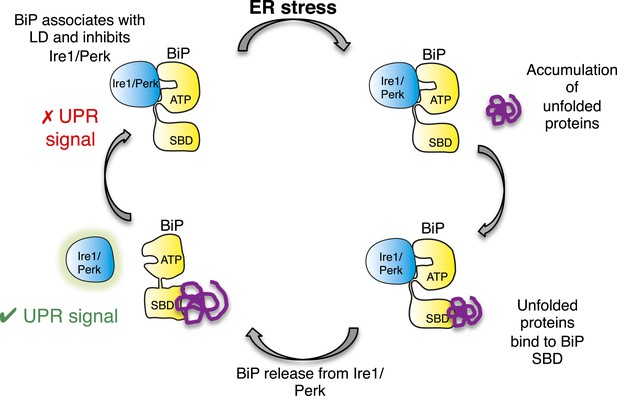
Allosteric model of UPR induction.
In the absence of misfolded protein, BiP interacts with UPR luminal domains, which acts to repress the UPR signal. Upon ER stress, unfolded protein binds to the canonical BiP substrate binding domain, which in turn causes the noncanonical BiP ATPase-luminal domain interaction to dissociate, ultimately leading to UPR signal activation/propagation.
Tables
Construct sizes for all BiP, Ire1 and Perk in vitro constructs used in this study
| Protein | Residue range |
|---|---|
| BiP FL | 28–654 |
| BiP ATPase | 28–405 |
| BiP SBD | 422–654 |
| Ire1 I–V | 24–440 |
| Ire1 I–IV | 24–390 |
| Ire1 II–V | 32–440 |
| Ire1 II–IV | 32–390 |
| Ire1 LD for cross-link experiment | 32–390 |
| Perk I–V | 54–509 |
| Perk I–IV | 54–403 |
| Perk II–V | 105–509 |
| Perk II–IV | 54–403 |





