B4GAT1 is the priming enzyme for the LARGE-dependent functional glycosylation of α-dystroglycan
Figures
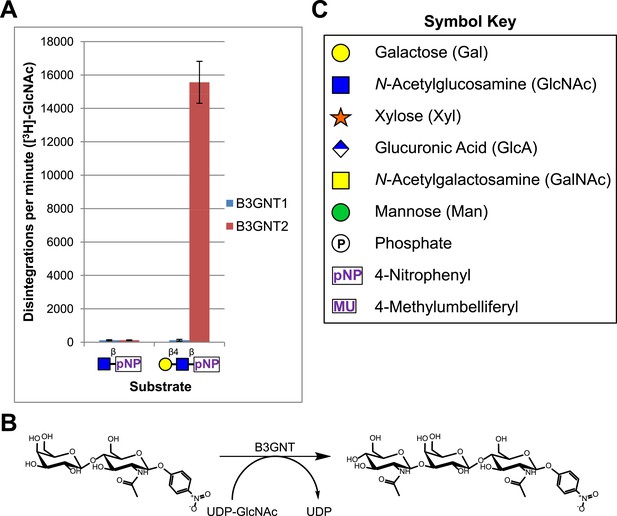
B3GNT1 does not possess β-1,3-N-acetylglucosaminyltransferase activity.
(A) The β-1,3-N-acetylglucosaminyltransferase activity of B3GNT1 towards pNP tagged N-acetyllactosamine was compared to that of B3GNT2 (reaction scheme presented in (B)). Incubations were carried out overnight at 37°C in 0.1 M MES pH 6.5 containing 10 mM MnCl2, 40,000 DPM UDP-[3H]GlcNAc and 2 mM non-radioactive UDP-GlcNAc. pNP-sugars were isolated from sugar nucleotide by reverse-phase C18 spin columns. UDP-[3H]GlcNAc transfer to Gal-β4-GlcNAc-β-pNP was measured by liquid scintillation counting. Averaged results from three independent experiments with error bars indicating standard deviation are shown. GlcNAc-β-pNP was used as a negative control acceptor. (C) Relevant sugar code symbols from ‘Essentials of Glycobiology’ as well as other symbols used throughout the paper are shown.
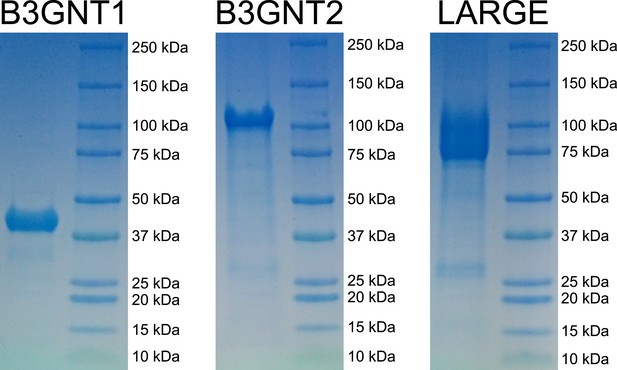
Coomassie brilliant blue G-250 stained SDS-PAGE gels of purified enzyme samples.
The prominent bands appear at the appropriate molecular weights for our constructs. B3GNT1 was cleaved from tag leaving only the catalytic domain while the other enzymes were not cleaved from expression construct elements (affinity tag and GFP).

Gal-β4-GlcNAc-β-pNP synthesized using bovine B4GALT1 LC-MS and LC-MS/MS spectra of Gal-β4-GlcNAc-β-pNP produced using B4GALT1.
Low m/z fragments include diagnostic GlcNAc oxonium ions at 186 and 168.
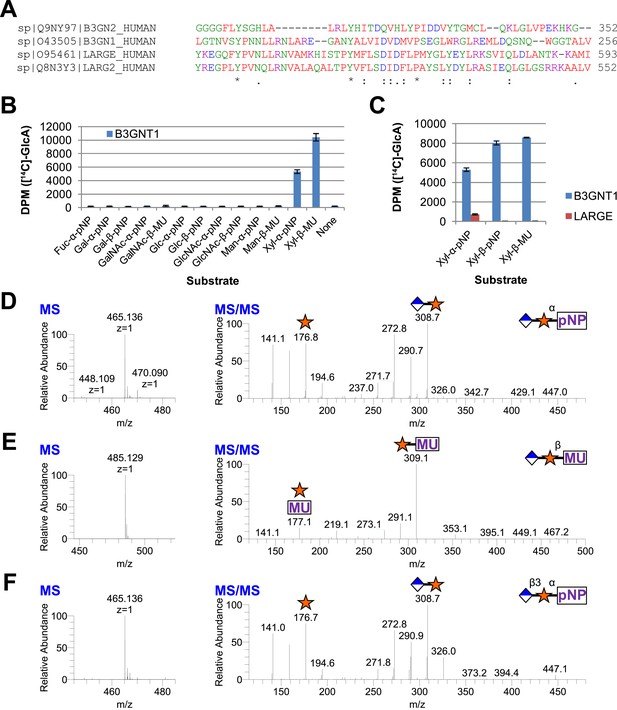
B3GNT1 is a glucuronyltransferase that uses xylose as an acceptor.
B3GNT1 is in CAZy family GT49 along with LARGE and LARGE2. (A) Clustal Omega multiple sequence alignment excerpt showing the strong alignment of B3GNT1 with the glucuronyltransferase domain of LARGE and LARGE2, including the DXD motif shown to be important for activity. B3GNT2 does not align well with the other inputs. (B) UDP-[14C]GlcA transfer screen assayed by liquid scintillation counting (disintegrations per minute–DPM), averaged results of three independent experiments with error bars indicating standard deviation. B3GNT1 transfers to xylose in both anomeric configurations. (C) LARGE transfers only to α-linked xylose as previously reported. (D) LC-MS and LC-MS/MS data showing B3GNT1 transfer to Xyl-α-pNP. The ammonium adduct is the dominant species (465.136) however the protonated species is observable at 448.109. The majority of unlabeled fragment peaks represent losses of water from labeled peaks. (E) LC-MS and LC-MS/MS data showing B3GNT1 transfer of UDP-GlcA to Xyl-β-MU. (F) LC-MS and LC-MS/MS data showing LARGE transfer to Xyl-α-pNP. Reactions were carried out overnight in 0.1 M MES pH 6.5 containing 10 mM MnCl2, 5 mM MgCl2, 2 mM substrate, 40,000 DPM UDP-[14C]GlcA and 2 mM non-radioactive donor.
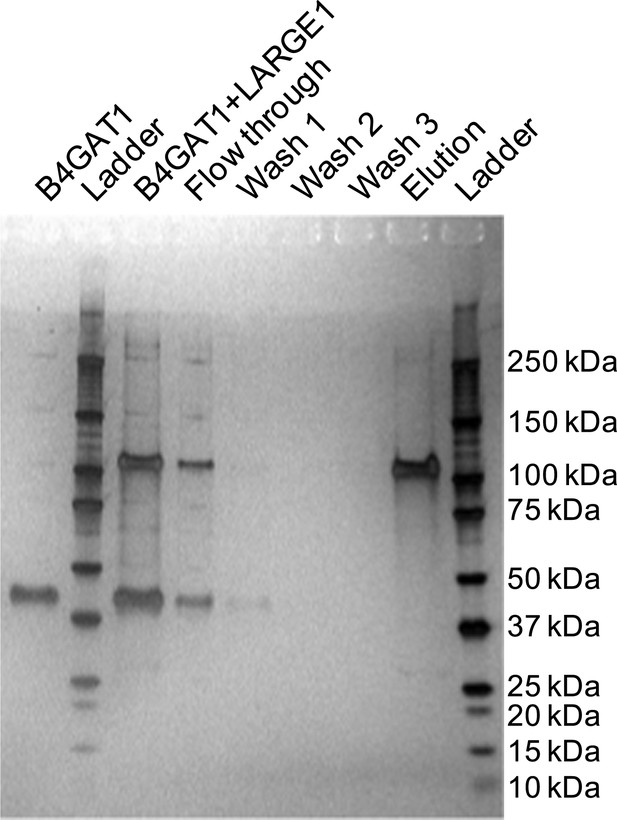
Silver stained SDS-PAGE gel of complex formation assay samples.
B4GAT1 (lacking an 8xHis tag) and LARGE1 (fused to an 8xHis tag) were mixed together and incubated for 15 min at 4°C. After incubation, the mixture was bound to Ni-NTA resin, washed three times with 20 mM imidazole containing PBS pH 7 and eluted in 250 mM imidazole in PBS pH 7. Aliquots were run by SDS-PAGE and silver stained. Evidence for in vitro complex formation was not observed as is seen by the lack of a B4GAT1 band in the elution fraction.
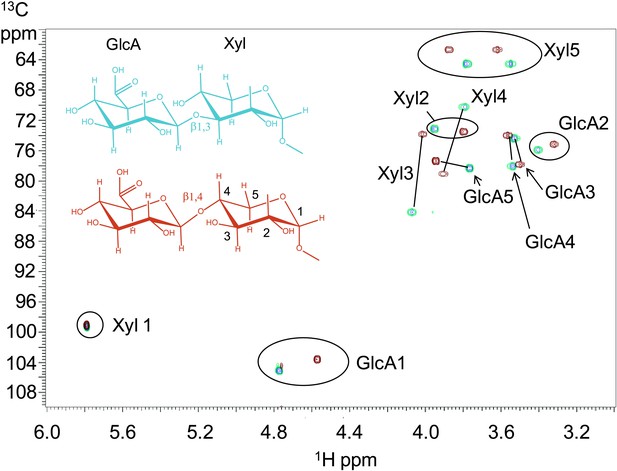
B3GNT1 is a β1,4-glucuronyltransferase in contrast to LARGE which is a β1,3-glucuronyltransferase.
1H–13C HSQC spectrum with peak assignments and structures of the products resulting from the action of B4GAT1 and UDP-GlcA on Xyl-α-pNP (red) and LARGE and UDP-GlcA on the same substrate (blue). Cross peaks are at the intersection of the 13C and 1H shifts of the partners in each C-H pair. The changes in the spectrum between the two compounds reflect the differences in the site of the glycosidic linkage formed, and particularly the major differences in the carbon shifts for the Xyl 3 and Xyl 4 sites between the two disaccharides is diagnostic of the change in their participation in the two different respective glycosidic linkages. The β configuration of the GlcA 1 site is confirmed by the 1-bond 1H–13C couplings of 166 Hz for these sites, as well as couplings between the H1 and H2 protons, 7.9–8.0 Hz. (data not shown).
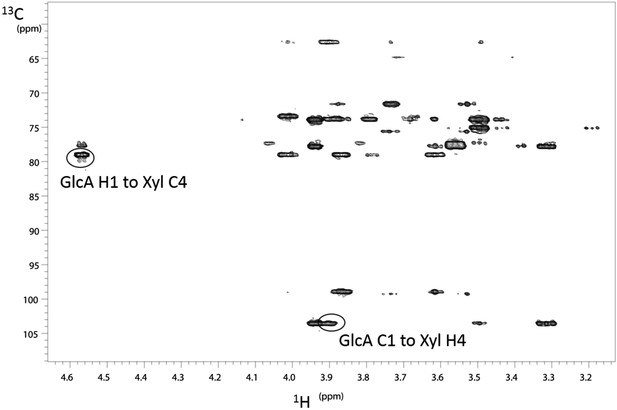
NMR determination of the 1,4 glycosidic linkage.
Section of the 1H–13C HMBC spectrum of the B4GAT1 product of GlcA addition to Xyl-α-pNP with both of the key through bond 1H–13C connections across the glycosidic linkage highlighted, unequivocally identifying the 1,4 glycosidic linkage.
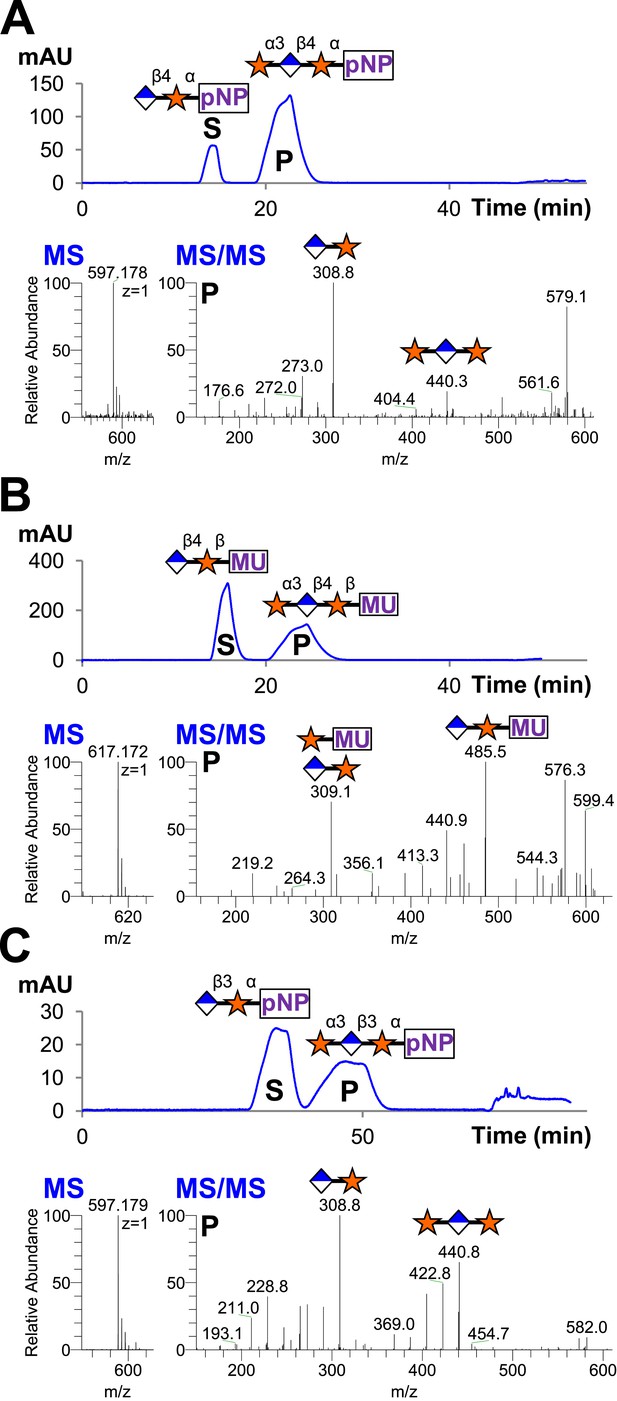
B4GAT1 disaccharide products are substrates for the xylosyltransferase activity of LARGE.
S indicates substrate peaks, P indicates product peaks (xylose added). Separation was carried out by isocratic C18 reverse-phase HPLC with absorbance monitoring at 310 nm. Products were confirmed by accurate mass and MS/MS fragmentation shown below with assignment of key characteristic peaks. (A) P = Xyl-α3-GlcA-β4-Xyl-α-pNP. (B) P = Xyl-α3-GlcA-β4-Xyl-β-MU. (C) The product of the sequential dual enzymatic activity of LARGE as a positive control; P = Xyl-α3-GlcA-β3-Xyl-α-pNP. Reactions were carried out overnight in 0.1 M MES pH 6.5 containing 10 mM MnCl2, 5 mM MgCl2, 2 mM substrate and 2 mM UDP-Xyl.
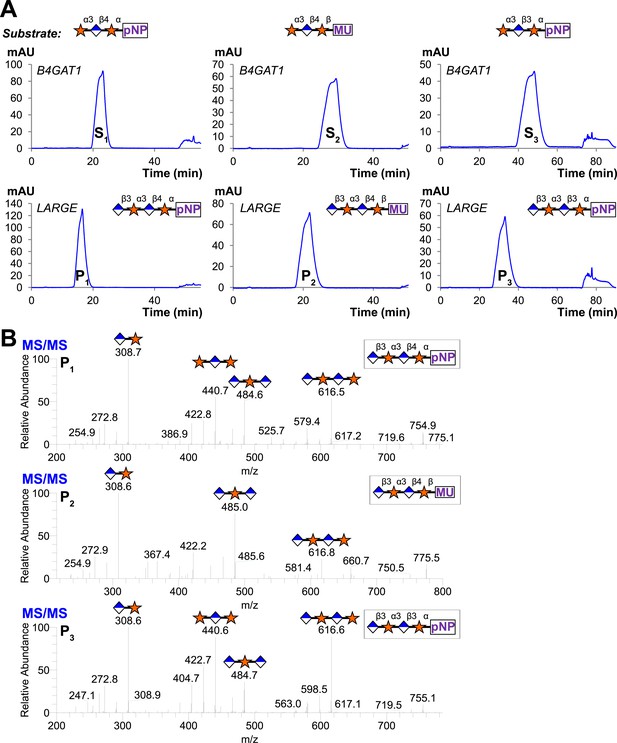
Trisaccharides terminating in an a3-xyloside are inefficient substrates for B4GAT1 in contrast to LARGE.
Purified trisaccharides were incubated with LARGE or B4GAT1 plus UDP-GlcA overnight and then separated by isocratic reverse-phase C18 HPLC. (A) P with a subscript indicates product (addition of GlcA) whereas S with a subscript indicates an unmodified trisaccharide substrate. The top chromatogram in each pair shows the result of incubation with B4GAT1, the bottom shows the result of incubation with LARGE. (B) Results were confirmed by MS/MS fragmentation spectra, only product spectra are shown. Reactions were carried out overnight in 0.1 M MES pH 6.5 containing 10 mM MnCl2, 5 mM MgCl2, 2 mM substrate and 2 mM UDP-GlcA.
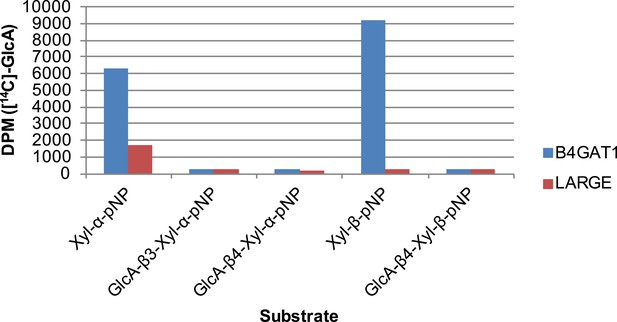
B4GAT1 and LARGE do not appear to possess branching activity.
Using purified disaccharide products of B4GAT1 or LARGE, we investigated B4GAT1 and LARGE for potential branching activity. Xyl-α-pNP and Xyl-β-pNP were included as controls. Transfer of GlcA by B4GAT1 or LARGE to disaccharides containing a terminal GlcA residue was not detected by radioactive transfer assay. Reactions were carried out overnight in 0.1 M MES pH 6.5 containing 10 mM MnCl2, 5 mM MgCl2, 2 mM substrate, 40,000 DPM UDP-[14C]GlcA and 2 mM non-radioactive donor.
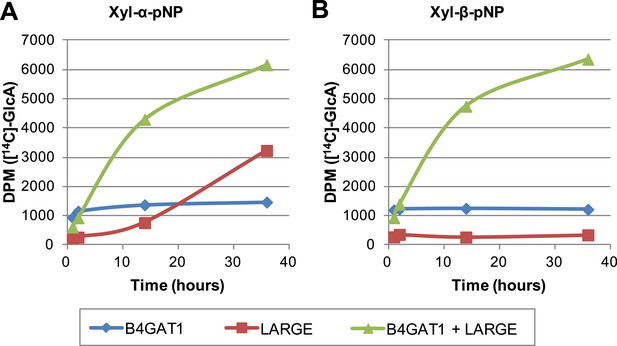
The slow reaction velocity of LARGE is rescued by addition of B4GAT1.
Reactions were carried out in 0.1 M MES pH 6.5 with 10 mM MnCl2, 5 mM MgCl2, 40,000 DPM UDP-[14C]GlcA and 10 mM non-radioactive UDP-GlcA and UDP-Xyl each. Aliquots were removed at the displayed time points, boiled, and processed using RP C18 spin columns to separate untransferred donor from substrate. (A) The initial transfer of GlcA by LARGE to Xyl-α-pNP is slow in the absence of B4GAT1 but after transfer of the first GlcA polymerization rates increase to mirror those of LARGE in the presence of B4GAT1. (B) With Xyl-β-pNP as substrate, B4GAT1, which only possesses glucuronyltransferase activity, transfers a single GlcA per molecule of substrate. LARGE is unable to transfer GlcA to Xyl-β-pNP. Polymerization is only observed with the addition of both enzymes to the reaction mixture.

B4GAT1 and LARGE combined produce extended GlcA-Xyl polymer chains.
B4GAT1 and LARGE were incubated together for 36 hr in 0.1 M MES pH 6.5 containing 10 mM MnCl2, 5 mM MgCl2, 10 mM UDP-GlcA, 10 mM UDP-Xyl and 1 mM Xyl-α-pNP. Mass spectra were acquired on a Bruker microflex and depict polymer formation. Note, due to likely decreased ionization efficiency with increasing polymerization, the sizes of observed peaks are unlikely to represent true abundances of each polymer species in the mixture.
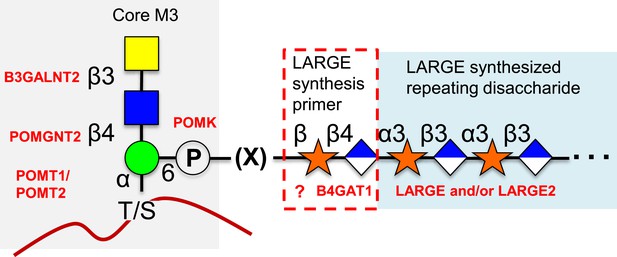
Proposed role of B4GAT1 in the O-mannosylation pathway.
The molecular link between the phospho-O-mannose trisaccharide synthesized on α-dystroglycan and the LARGE synthesized repeating disaccharide crucial for laminin binding reactivity is still not fully characterized (represented as [X]). B4GAT1 appears to possess a priming activity for LARGE and likely adds to an underlying β-xylose that is added by an as yet undefined glycosyltransferase (represented with a question mark).
Tables
Kinetics at 2 mM UDP-GlcA
| Enzyme | Substrate | Km (mM) | kcat (s−1) | kcat/Km (M-1 s−1) | Specific activity (pmol/min/μg) |
|---|---|---|---|---|---|
| B3GNT1 | Xyl-α-pNP | 3.0 | 0.087 | 29 | 130 |
| Xyl-β-pNP | 4.0 | 0.25 | 63 | 380 | |
| LARGE | Xyl-α-pNP | 6.0 | 0.005 | 0.84 | 2.7 |
| Xyl-β-pNP | NM | NM | NM | NM |
-
NM = not measurable (below limit of detection).
Additional files
-
Supplementary file 1
Top 5 protein search results in each purified enzyme sample LC-MS/MS was carried out on tryptic digests of purified enzyme samples. Results filtered to peptide 1% false-discovery rate are displayed from a Proteome Discoverer 1.4 Sequest HT search against the Uniprot human database along with total score and number of peptide spectral matches for each protein assignment (# PSMs).
- https://doi.org/10.7554/eLife.03943.017
-
Supplementary file 2
Chemical shifts of disaccharides. Chemical shifts of the disaccharide portion of the products of LARGE and B4GAT1 addition of GlcA to Xyl-α-pNP. Proton shifts are referenced to the HDO signal, 4.77 ppm at 25°C relative to DSS, and 13C shifts relative to DSS at 0 ppm were then determined using indirect referencing to the proton standard.
- https://doi.org/10.7554/eLife.03943.018





