Phosphatidylinositol 3-kinase and COPII generate LC3 lipidation vesicles from the ER-Golgi intermediate compartment
Figures
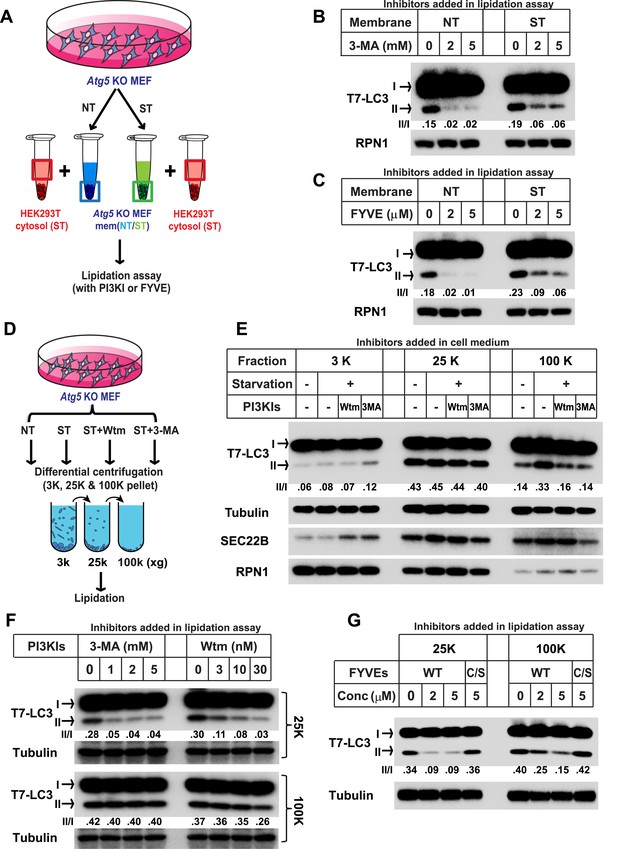
Starvation and PI3K-dependent generation of small membranes for LC3 lipidation.
(A–C) Atg5 KO MEFs were either untreated (NT) or starved (ST) with EBSS (Earle's Balanced Salt Solution) for 30 min. Total membranes (mem) from lysed cells were collected and incubated in a lipidation reaction with cytosols prepared from starved HEK293T cells. Reactions contained the indicated concentrations of PI3K inhibitor (PI3KI) 3-methyladenine (3-MA) (B) or FYVE protein (C). A diagram of the experimental scheme is shown in (A). RPN1, Ribophorin 1 (D, E) Atg5 KO MEFs were either untreated (NT) or starved (ST) with EBSS in the absence or presence of 20 nM wortmannin (Wtm) or 10 mM 3-methyladenine (3-MA) for 30 min. Membranes from each treated cell sample were collected and subjected to a differential centrifugation to separate the 3K ×g, 25K ×g and 100K ×g pellet fractions followed by a lipidation assay as above (E). A diagram is shown in (D). (F, G) Atg5 KO MEFs were starved for 30 min. Membranes in the 25K ×g and 100K ×g pellets from a differential centrifugation were collected as described above. A similar lipidation assay was performed in the presence of indicated concentrations (Conc in G) of 3-MA, wortmannin (F) and FYVE protein as well as a PI3P binding-deficient FYVE mutant protein (C/S) (G). Quantification of lipidation activity is shown as the ratio of LC3-II to LC3-I (II/I).
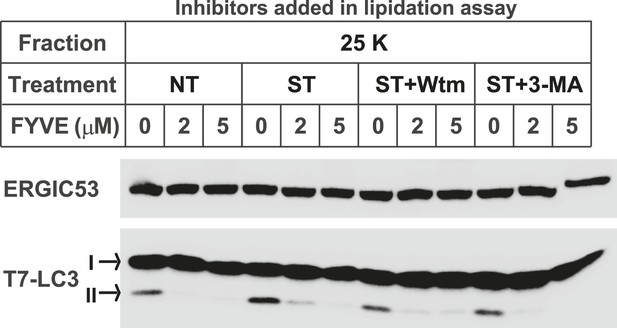
The FYVE domain protein blocks LC3 lipidation of the 25K membrane pellet fraction.
Atg5 KO MEFs were either untreated (NT) or starved (ST) with EBSS in the absence or presence of 20 nM wortmannin or 10 mM 3-MA for 30 min as shown in Figure 1D. The 25K membrane fractions were collected from lysed cells from each condition. LC3 lipidation was performed with the 25K membrane fractions in the presence of the indicated concentrations of FYVE domain protein.
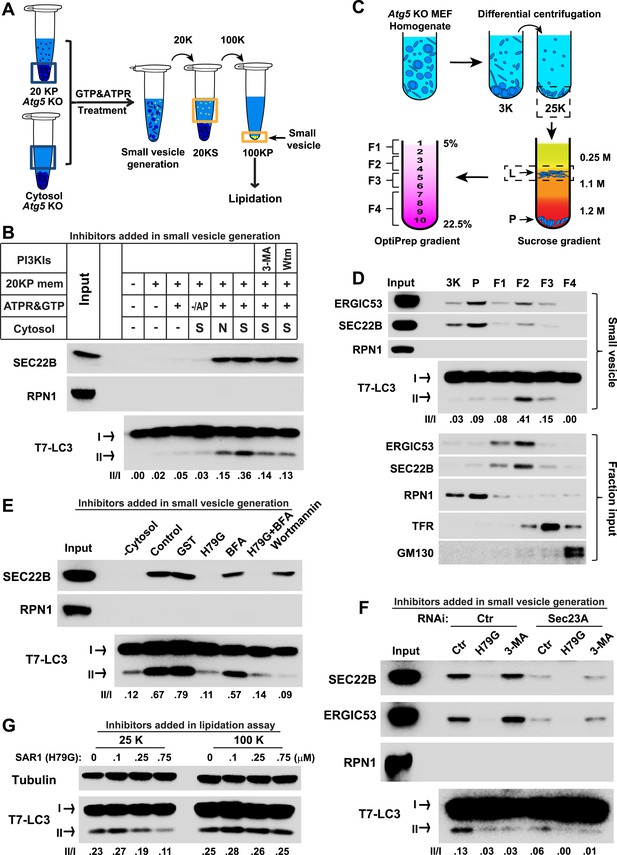
COPII and PI3K-dependent generation of small vesicles from ERGIC for LC3 lipidation.
(A) Diagram showing the cell-free system to generate small vesicles for LC3 lipidation. Briefly, a medium-speed membrane pellet (20K ×g, 20KP) from Atg5 KO MEFs was harvested and incubated with cytosol from Atg5 KO MEFs, GTP and an ATP regeneration system (ATPR), and in the absence or presence of PI3K inhibitors (Figure 2B) or the PI3P blocker FYVE domain protein (Figure 2—figure supplement 1). The slowly-sedimenting membranes generated were separated from the donor membrane by a medium speed centrifugation (20K ×g) and collected by high-speed sedimentation (100K ×g) of the supernatant fraction (20KS). A lipidation assay with HEK293T cytosol was performed to analyze the competency of the small membranes (100KP) to induce LC3 lipidation. (B) A small vesicle generation assay as above was performed with the indicated conditions. Slowly-sedimenting vesicles were collected to determine activity in the LC3 lipidation reaction. 3-MA, 5 mM; Wortmannin, 20 nM; S, starved Atg5 KO MEF cytosol; N, untreated Atg5 KO MEF cytosol; −/AP, in the absence of ATP regeneration and GTP and in the presence of apyrase (AP). (C) Diagram showing the fractionation procedure and fractions collected for the small vesicle generation assay shown in (D). Briefly, Atg5 KO MEF membranes were subjected to differential centrifugation to collect 3K ×g and 25K ×g pellet fractions. Sucrose gradient ultracentrifugation was performed to separate the L and P fraction after which the L was further separated by OptiPrep gradient ultracentrifugtion. Ten fractions from top to bottom were collected and combined to four as indicated (F1-4). (D) A small vesicle generation assay followed by a lipidation reaction was performed with the 3K, P and F1-4 fractions (upper four panels: Small vesicle). SDS-PAGE and immunoblot with indicated antibodies were carried out to probe the organelle enrichment of each fraction (lower five panels: Fraction input). TFR, transferrin receptor (E) A small vesicle generation assay as shown in (A) was performed in the presence of indicated proteins or drugs. Slowly-sedimenting vesicles were collected and incubated in a lipidation reaction in the presence of GST, H79G (Sar1A (H79G), 0.7 μM), wortmannin (20 nM), or BFA (brefeldin A, 0.5 μg/ml). (F) A small vesicle generation assay as shown in (A) was performed with cytosols from starved Atg5 KO MEF from control or Sec23A knockdown cells in the absence or presence of Sar1A (H79G) or 3-MA followed by a lipidation reaction. (G) Atg5 KO MEFs were starved for 30 min. Differential centrifugation as shown in Figure 1D was performed to collect the 25K ×g and 100K ×g pellet fractions. The LC3 lipidation was performed on these two fractions with indicated concentrations of SAR1 (H79G). Quantification of lipidation activity is shown as the ratio of LC3-II to LC3-I (II/I).
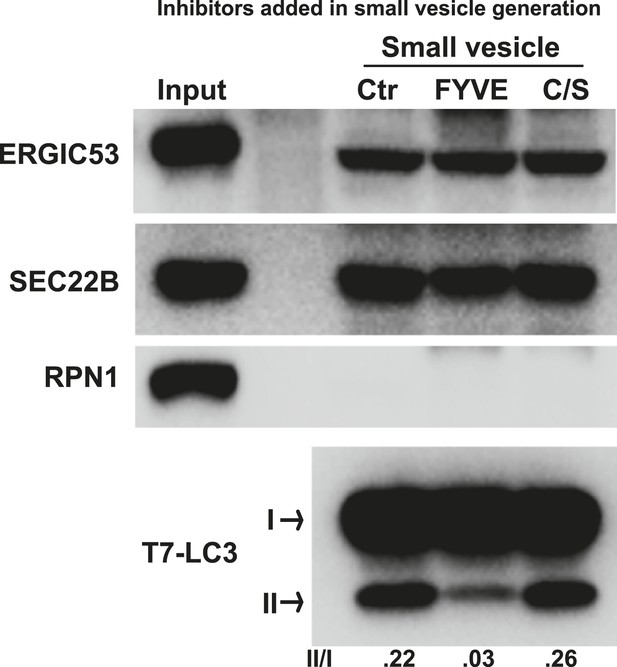
PI3P is required for generation of the LC3 lipidation-active vesicles in vitro.
A small vesicle generation assay as shown in Figure 2A was performed in the absence or presence of indicated FYVE domain proteins. Vesicles were then incubated in the LC3 lipidation reaction.
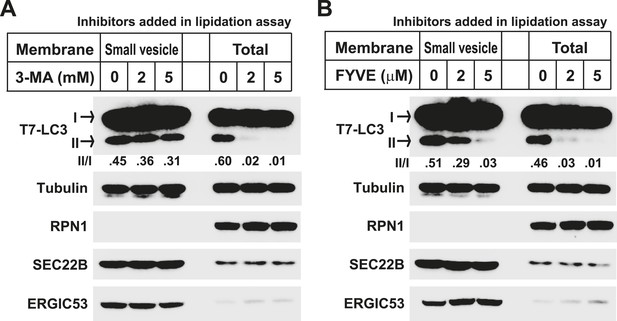
Lipidation on the small vesicles generated in vitro is resistant to PI3K inhibition.
(A, B) A small vesicle generation assay as shown in Figure 2A was performed. Small vesicles as well as the total membranes were collected and subjected to the LC3 lipidation assay in the presence of indicated concentrations of 3-MA (A) or FYVE domain protein (B).
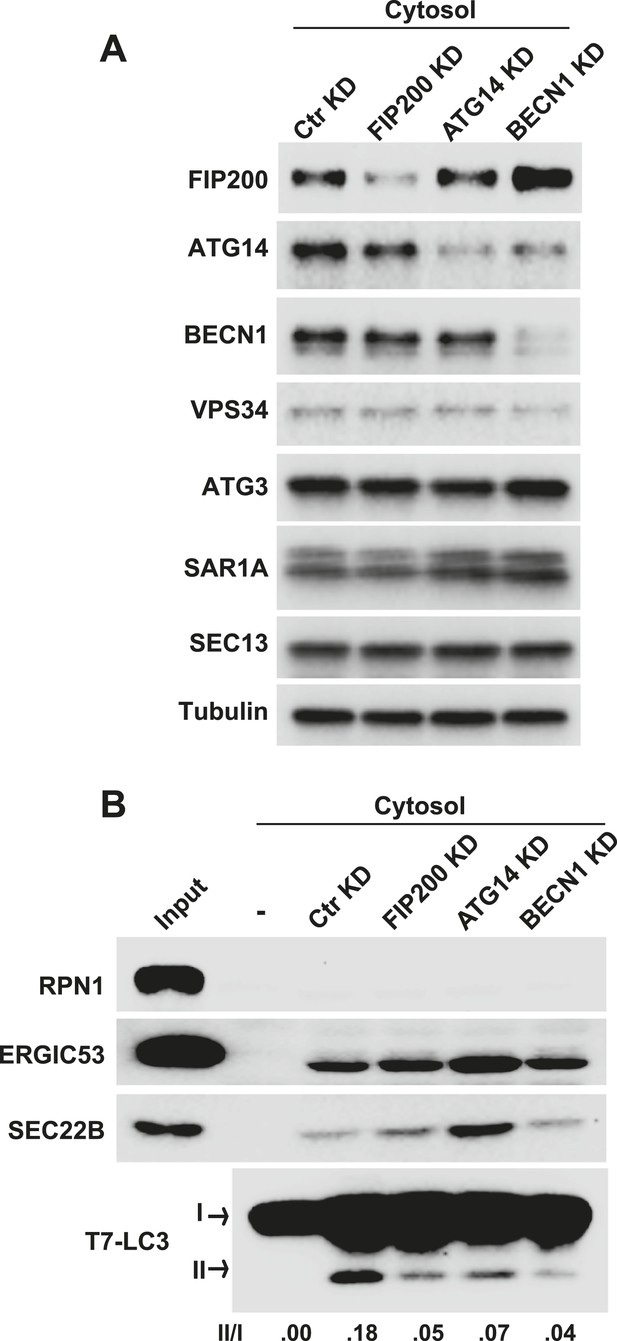
Early autophagic factors are required for the generation of small vesicles active in LC3 lipidation.
(A, B) Atg5 KO MEFs were transfected with indicated siRNAs. At 72 hr after transfection, cytosol was extracted from the cells followed by a small vesicle generation assay with Atg5 KO MEF membrane as shown in Figure 2A (B). RNAi efficiency was determined by immunoblot with indicated antibodies (A). BECN1, Beclin-1.
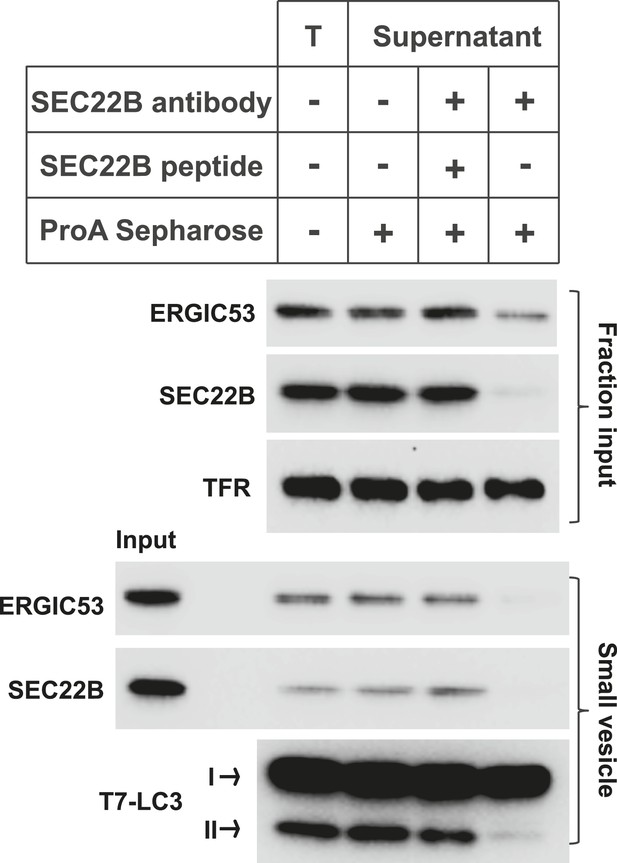
Immunodepletion of the ERGIC abolishes the generation of LC3 lipidation-active vesicles.
The L fraction as shown in Figure 2C was collected and were incubated without or with SEC22B antibody (Ab), or with SEC22B antibody together with the blocking peptide (SEC22B pep). Protein A Sepharose was added to pull down SEC22B antibody as well as the associated membranes. Then the membranes from the supernatant were collected and the small vesicle generation assay was performed with these membranes followed by the LC3 lipidation assay. T, total L fraction before depletion; TFR, transferrin receptor.
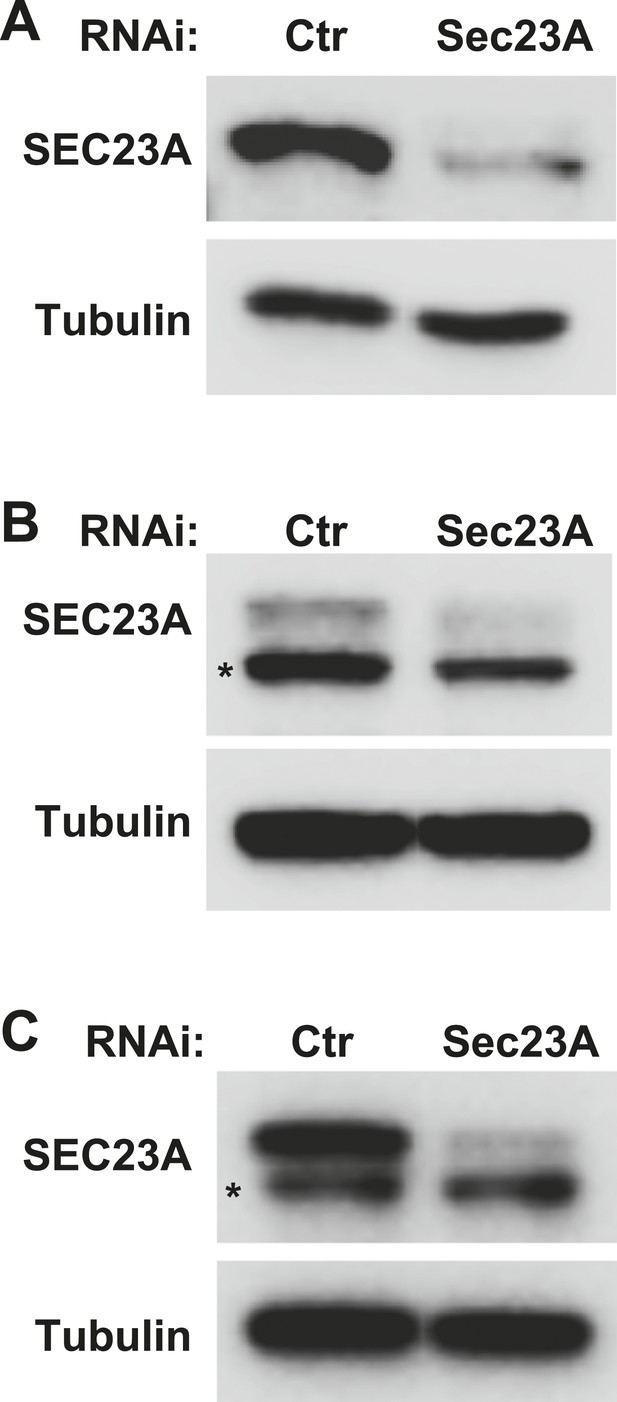
Knockdown efficiency of the Sec23A siRNA.
(A–C) Atg5 KO (A), WT (B) MEFs and Hela cells (C) were transfected with indicated siRNAs. After transfection (72 hr), cells were starved for 1 hr followed by lysis to collect cytosol fractions. SDS-PAGE and immunoblot were performed to determine knockdown efficiency. Asterisk, non-specific band.
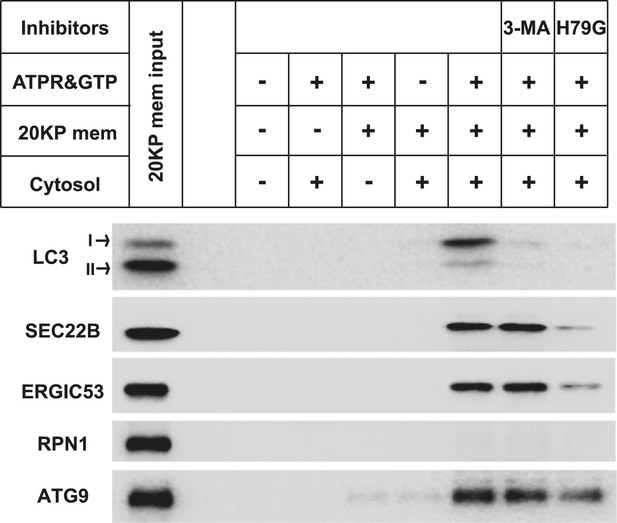
PI3K and COPII-dependent generation of small vesicles positive for endogenous LC3 with WT MEF cytosol and membrane.
A small vesicle generation assay as shown in Figure 2A with WT MEF cells was performed. In brief, the 20KP membrane of untreated WT MEF and cytosol from starved WT MEF were collected and incubated with an ATP regeneration system, GTP, the PI3K inhibitor 3-MA and COPII budding inhibitor Sar1 (H79G) in combinations indicated in the figure. After 1 hr incubation at 30°C, the small vesicles generated were collected by the differential centrifugation as shown in Figure 1A. Immunoblot was performed with indicated antibodies.
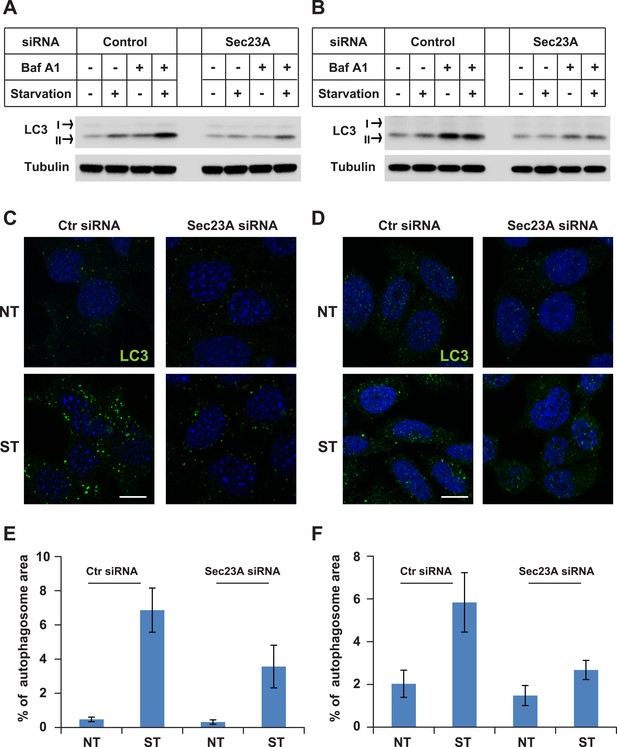
Knockdown of Sec23A inhibits autophagosome biogenesis.
(A, B) WT MEFs (A) or Hela cells (B) were either untreated or starved with or without bafilomycin A1 (200 ng/ml) for 40 min. Then the cells were lysed by SDS-PAGE buffer followed by immunoblot. (C, D) WT MEFs (C) or Hela cells (D) were either untreated or starved for 1 hr. Immunofluorescence was performed with LC3 antibody. Bar: 10 μM (E, F) Quantification of autophagosome biogenesis as shown in (C, D) based on the percentage of LC3 puncta area to the cell area. Error bars represent standard deviations from 10 images (∼50 cells) in three independent experiments.
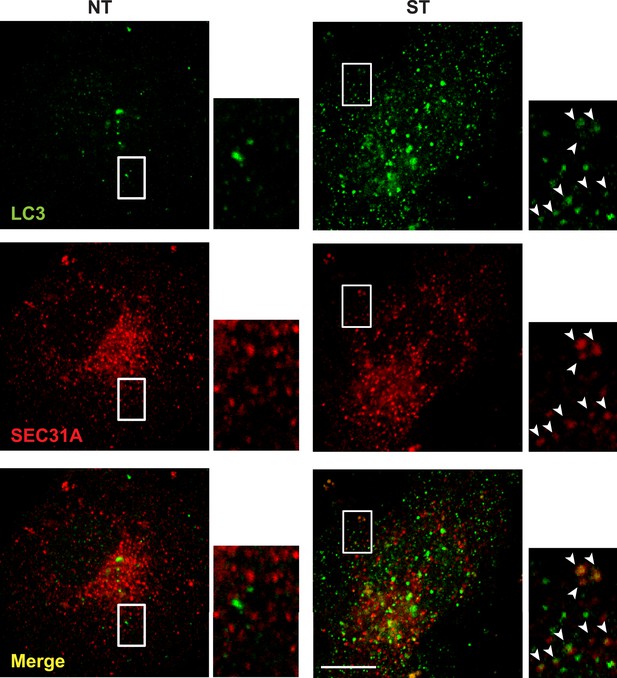
Small LC3 vesicles colocalize with COPII after starvation COS7.
cells were transfected with a plasmid encoding an HA-tagged SEC31A. 24 hr after transfection, the cells were either untreated (NT) or starved (ST) with EBSS for 1 hr. Then immunofluorescence was performed with LC3 antibody and HA antibody to label endogenous LC3 and the expressed HA-SEC31A. Arrowheads indicate the colocalized vesicles. Bar: 10 μM.
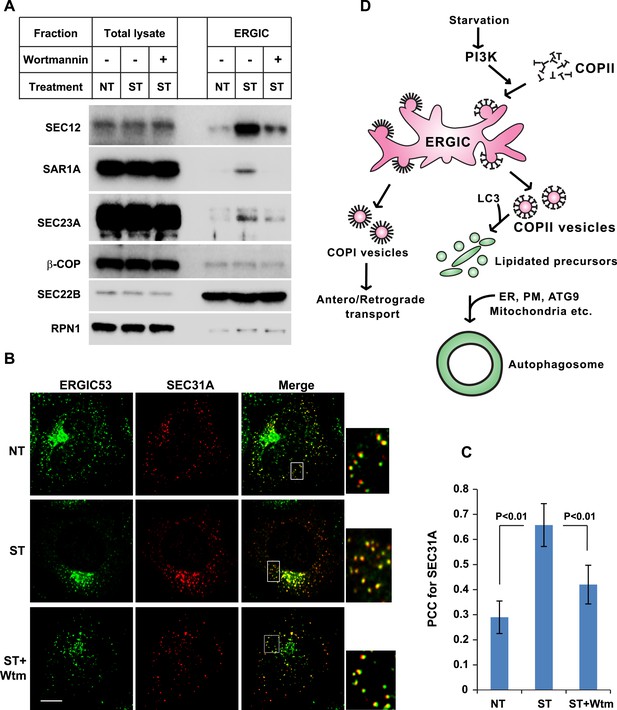
PI3K-dependent recruitment of COPII to ERGIC after starvation.
(A) Atg5 KO MEFs were either untreated or starved with or without 20 nM wortmannin for 30 min. Cells were harvested and ERGIC membranes were isolated by pooling fractions 3 and 4 (F2 in Figure 2C,D) of the OptiPrep gradient in the three-step fractionation approach followed by immunoblot to examine the amount of indicated markers on ERGIC membranes. (B) Atg5 KO MEFs were transfected with a plasmid encoding an HA-tagged SEC31A. After transfection (24 hr), the cells were treated as shown in (A). Immunofluorescence was performed with anti-ERGIC53 and anti-HA antibodies and the cells were examined by confocal microscopy. Bar: 10 μM (C) Quantification of SEC31A overlapped with ERGIC53 shown in (B) using the Pearson's correlation coefficient (PCC). Error bars represent standard deviations from ∼20 cells in three independent experiments. p values were calculated by T-test. (D) A proposed model. Starvation activates autophagic PI3K to induce COPII recruitment on ERGIC. COPII vesicles generated from ERGIC serve as templates for efficient LC3 lipidation. These membrane precursors may collaborate with other membranes such as the endoplasmic reticulum (ER), plasma membrane (PM), ATG9 compartment and mitochondria, to form a mature autophagosome.
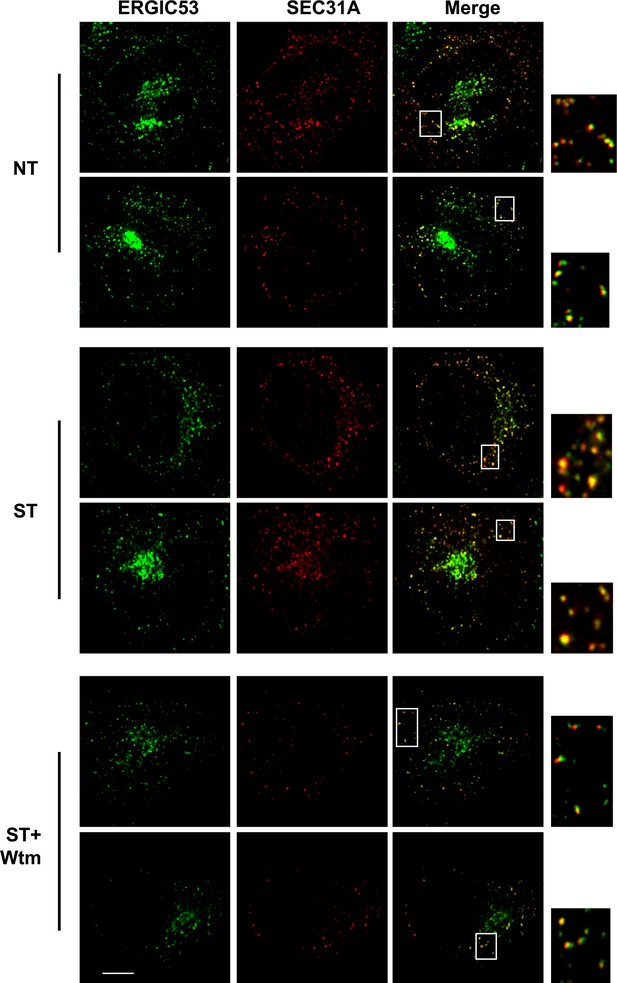
Extra images of Figure 3B.
Bar: 10 μM.
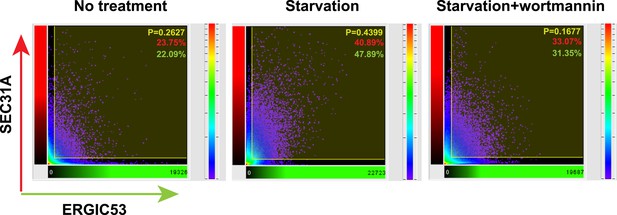
Histogram of colocalization between ERGIC53 and SEC31A in the deconvoluted 3D image.
Colocalization histogram of the original image was directly generated using Imaris imaging software. Percentage of colocalization between SEC31A (red) and ERGIC53 (green) as well as the Pearson's correlation coefficient (P) above the threshold was calculated by the software.
Videos
3D deconvolution images showing the distribution of ERGIC and COPII under conditions of untreated, starved or starvation with the PI3K inhibitor.
Atg5 KO MEFs were transfected with a plasmid encoding an HA-tagged SEC31A. Immunofluorescence labeling of ERGIC53 (green) and SEC31A (red) was performed as shown in Figure 3B. Z-stack images were collected and deconvolution was performed. 3D images were generated using Imaris imaging software. Cells with conditions of no treatment, starvation and starvation with 20 nM wortmannin were sequentially displayed.





