Reconstitution of bacterial autotransporter assembly using purified components
Figures
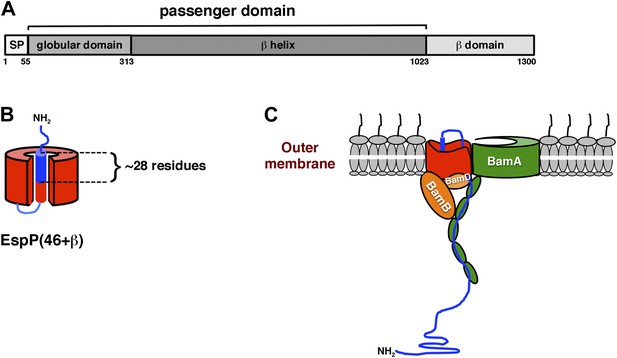
Domain structure of EspP and model for passenger domain translocation.
(A) EspP consists of a signal peptide (SP; residues 1–55), an extracellular (‘passenger’) domain (residues 56–1023) and a β barrel (‘β’) domain (residues 1024–1300) (Brunder et al., 1997). While most of the passenger domain forms a long β helix, the N-terminus (residues 56–313) forms a discrete globular domain (Khan et al., 2011). (B) Illustration of EspP(46+β). Prior to the release of the passenger domain in an intrabarrel cleavage reaction, ∼28 residues of the 46 residue passenger domain are embedded inside the β domain pore. (C) Available evidence indicates that the EspP passenger domain is secreted in a hairpin conformation while distinct regions of the β domain interact with BamA, BamB and BamD (Ieva et al., 2011). Components of the transport channel likely include the open β domain and/or the BamA β barrel, which has been proposed to open laterally (Noinaj et al., 2013, 2014). In any case, the finding that proteolytic maturation and the release of the β domain from the Bam complex both require the completion of translocation (Peterson et al., 2010; Ieva et al., 2011; Pavlova et al., 2013) strongly suggests that the active site cannot form during the passage of the passenger domain across the OM. BamC and BamE hve been omitted from the model for the sake of clarity.
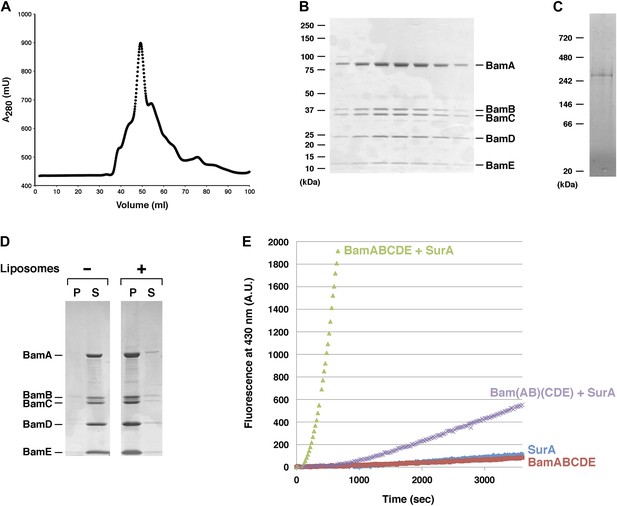
Purification and functional test of BamABCDE.
(A) Chromatogram of BamABCDE on S-200 gel filtration column. (B) SDS-PAGE analysis of the peak fractions in (A). Proteins were visualized by Coomassie Blue staining. (C) The peak fractions in (B) were pooled and analyzed by Blue Native PAGE. (D) Purified BamABCDE was centrifuged in the absence of liposomes or after reconstitution into liposomes. The pellet (P) and supernatant (S) fractions were analyzed by SDS-PAGE. (E) Urea denatured OmpT was diluted and incubated with SurA and proteoliposomes containing either BamABCDE (green) or Bam(AB)(CDE) (purple), proteoliposomes containing BamABCDE alone (red) or SurA alone (blue). OmpT activity was assessed by measuring the fluorescent signal generated by the cleavage of a fluorogenic peptide.
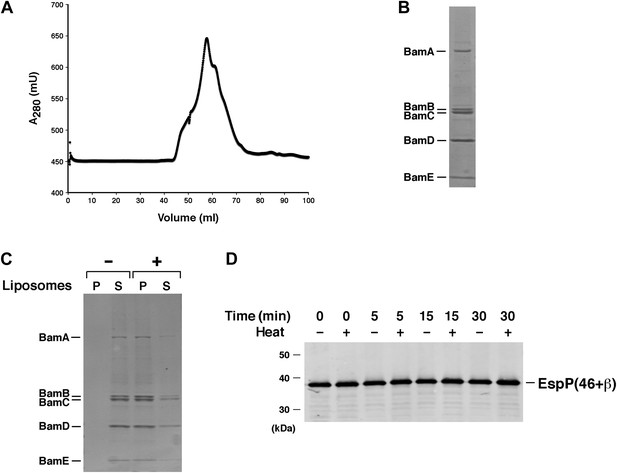
Test of purified Bam(AB)(CDE) in EspP assembly assay.
(A) Chromatogram of Bam(AB)(CDE) on S-200 gel filtration column. (B) The peak fractions in part A were pooled and analyzed by SDS-PAGE. Proteins were visualized by Coomassie Blue staining. (C) Purified Bam(AB)(CDE) was centrifuged in the absence of liposomes or after reconstitution into liposomes. The pellet (P) and supernatant (S) fractions were analyzed by SDS-PAGE. (D) Urea-denatured EspP(46+β) was incubated with SurA and proteoliposomes containing Bam(AB)(CDE). Aliquots were placed on ice at various time points, heated to 95°C or maintained at room temperature after the addition of SDS-PAGE buffer, and analyzed by Western blot using an anti-EspP C-terminal peptide antiserum.
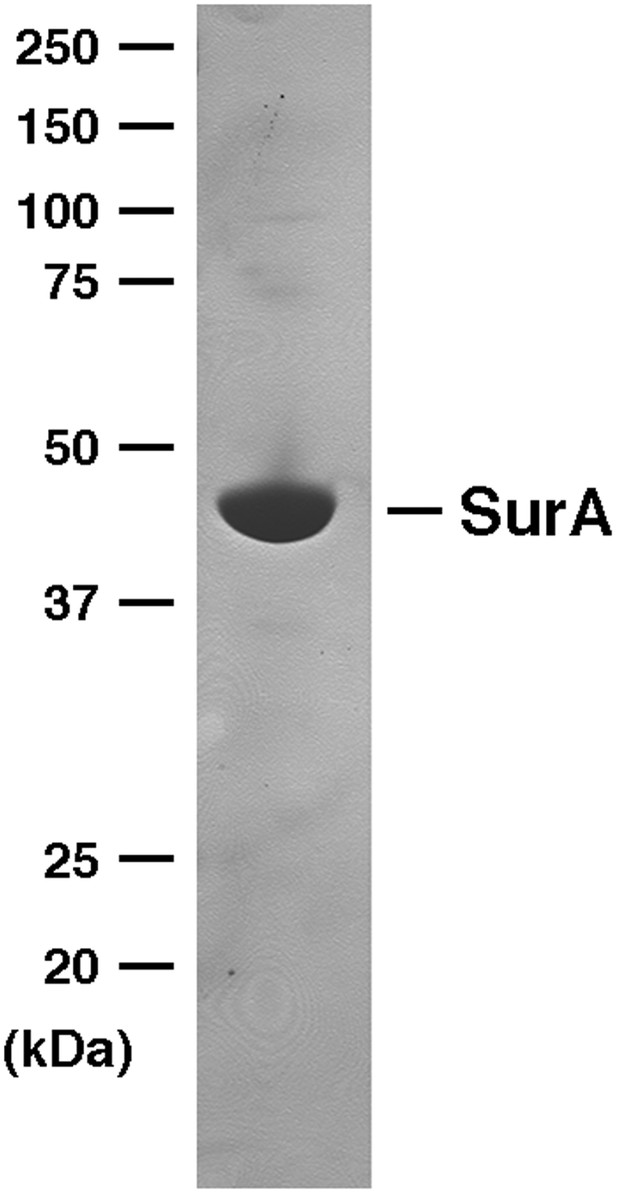
SDS-PAGE analysis of purified SurA.
His-tagged SurA was purified by affinity chromatography and analyzed by SDS-PAGE. The protein was visualized by Coomassie Blue staining.
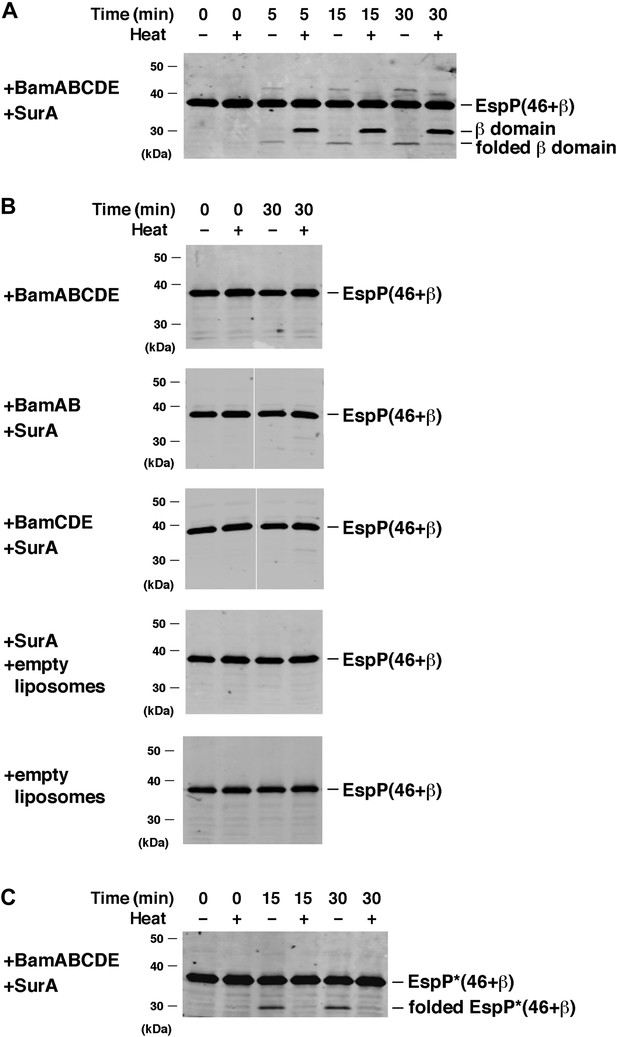
BamABCDE and SurA catalyze the assembly of EspP(46+β).
(A) Urea-denatured EspP(46+β) was incubated with SurA and proteoliposomes containing BamABCDE. Aliquots were placed on ice at various time points, heated to 95°C or maintained at room temperature after the addition of SDS-PAGE buffer, and analyzed by Western blot using an anti-EspP C-terminal peptide antiserum. (B) Urea-denatured EspP(46+β) was incubated with the indicated factors. Aliquots were removed after 0 and 30 min and analyzed as in (A). (C) Urea-denatured EspP*(46+β) was diluted and incubated with SurA and proteoliposomes containing BamABCDE. Aliquots were removed at various time points and analyzed as in (A).
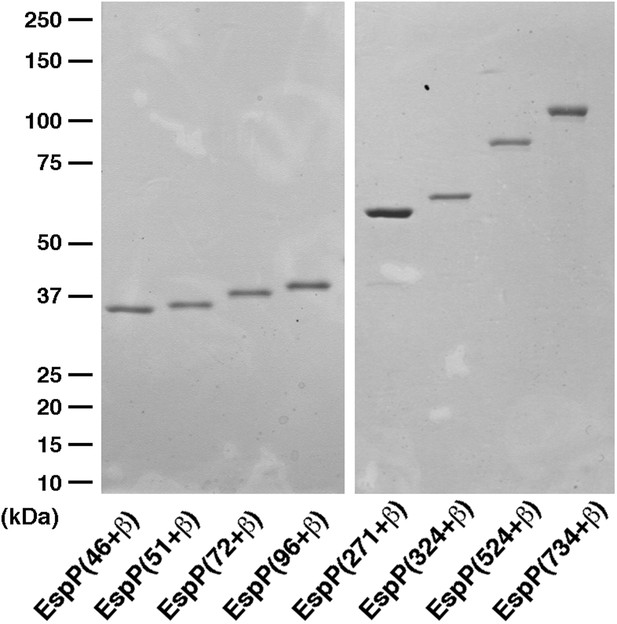
SDS-PAGE analysis of purified EspP derivatives.
The indicated EspP derivatives were purified from inclusion bodies and analyzed by SDS-PAGE. The proteins were visualized by Coomassie Blue staining.
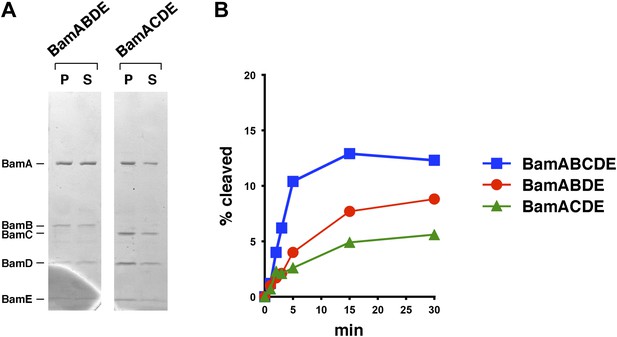
Bam complexes lacking BamB or BamC catalyze the assembly of EspP(46+β) less effectively than the holocomplex.
(A) Purified BamABDE and BamACDE were centrifuged after reconstitution into liposomes. The pellet (P) and supernatant (S) fractions were analyzed by SDS-PAGE. (B) Urea-denatured EspP(46+β) was incubated with SurA and proteoliposomes containing BamABCDE, BamABDE or BamACDE. Aliquots were placed on ice at various time points, heated to 95°C after the addition of SDS-PAGE buffer, and analyzed by Western blot using an anti-EspP C-terminal peptide antiserum. A quantitation of the data is shown.
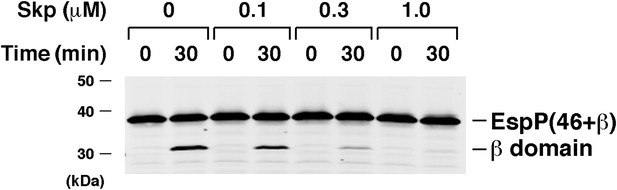
Skp inhibits the assembly of EspP(46+β).
Urea-denatured EspP(46+β) was incubated with SurA, proteoliposomes containing BamABCDE, and the indicated concentration of Skp. Aliquots were placed on ice after 0 and 30 min, heated to 95°C after the addition of SDS-PAGE buffer, and analyzed by Western blot using an anti-EspP C-terminal peptide antiserum.
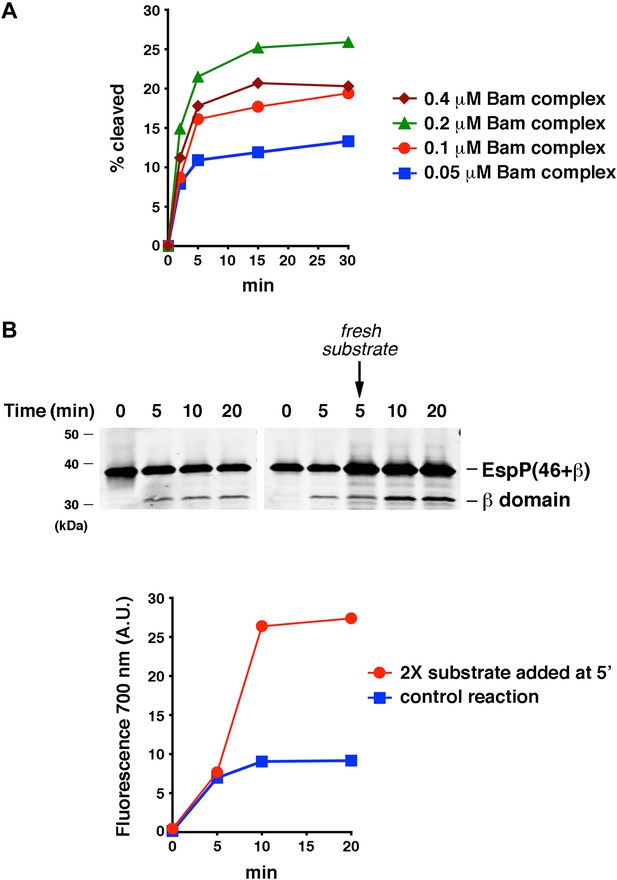
The efficiency of EspP(46+β) assembly is limited by the ability of the protein to remain folding-competent.
(A) Urea-denatured EspP(46+β) was incubated with SurA and proteoliposomes containing the indicated concentration of BamABCDE. Aliquots were placed on ice at various time points, heated to 95°C after the addition of SDS-PAGE buffer, and analyzed by Western blot using an anti-EspP C-terminal peptide antiserum. A quantitation of the data is shown. (B) Urea-denatured EspP(46+β) (0.1 μM) was mixed with SurA and proteoliposomes containing 0.2 μM BamABCDE, and the mixture was divided into two equal reactions. After 5 min fresh EspP(46+β) (0.2 μM) was added to one reaction. Aliquots from both reactions were placed on ice at various time points, heated to 95°C after the addition of SDS-PAGE buffer, and analyzed by Western blot using an anti-EspP C-terminal peptide antiserum. The graph shows a quantitation of the signal produced by the binding of a fluorescently labeled secondary antibody to the cleaved β domain on the Western blot.
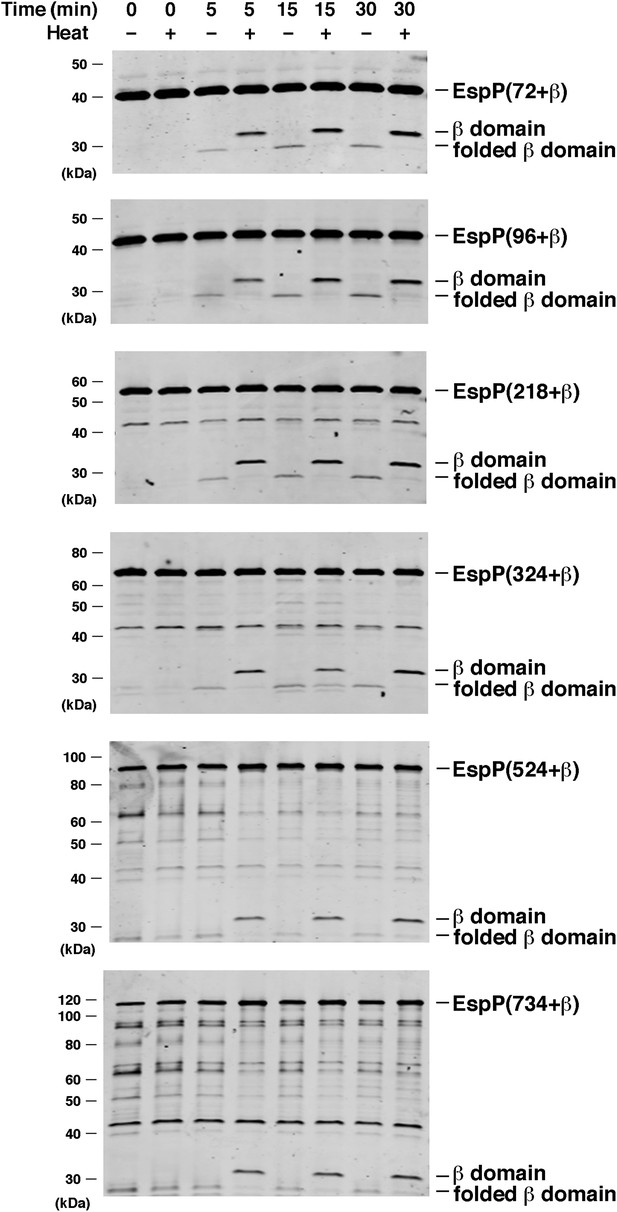
BamABCDE and SurA catalyze the assembly of longer EspP derivatives.
The indicated urea-denatured EspP derivative was incubated with SurA and proteoliposomes containing BamABCDE. Aliquots were removed at various time points, heated to 95°C or maintained at room temperature after the addition of SDS-PAGE buffer, and analyzed by Western blot using an anti-EspP C-terminal peptide antiserum. The ∼27 kDa polypeptide observed at the 0 min time point on the bottom two gels is an unidentified background band.
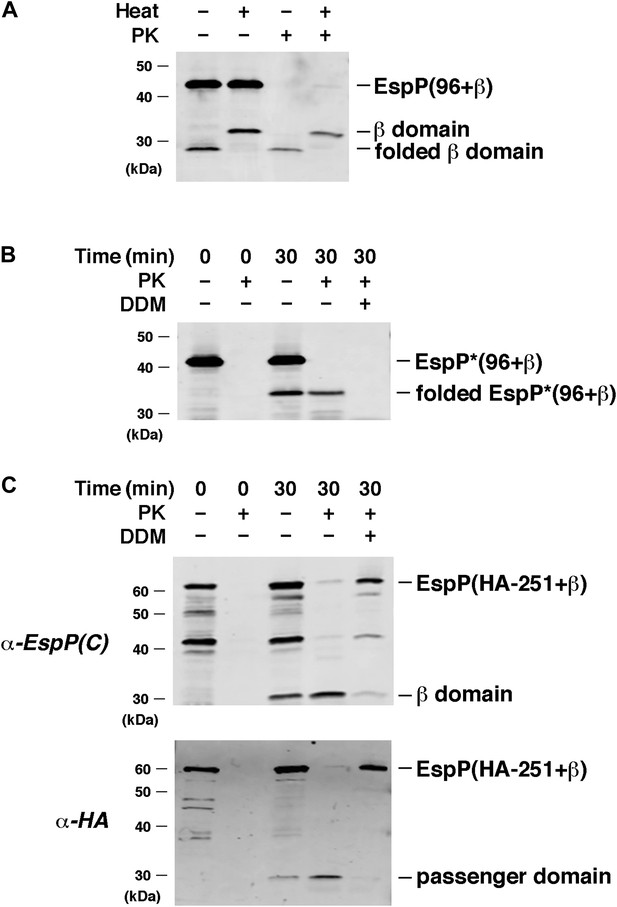
EspP derivatives are correctly assembled into proteoliposomes.
(A) Urea-denatured EspP(96+β) was incubated with SurA and proteoliposomes containing BamABCDE. Aliquots were placed on ice after 0 and 30 min and either treated with PK or left untreated. After the addition of SDS-PAGE buffer samples were heated to 95°C or maintained at room temperature and analyzed by Western blot using an anti-EspP C-terminal peptide. (B) The experiment described in (A) was repeated with EspP*(96+β), except that n-dodecyl β-D-maltoside (DDM) was added to one sample prior to PK treatment, and none of the samples were heated after the addition of SDS-PAGE buffer. (C) The experiment described in (A) was repeated with EspP(HA-251+β), except that DDM was added to one sample prior to PK treatment, and all of the samples were heated to 95°C after the addition of SDS-PAGE buffer. Samples were divided in half and analyzed by Western blot using an anti-EspP C-terminal peptide antiserum or an anti-HA antiserum.
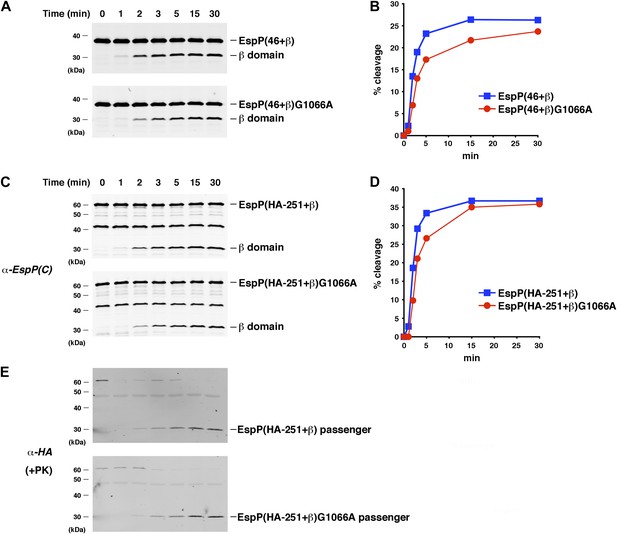
The assembly of the β domain is the rate-limiting step in EspP biogenesis.
(A) Urea-denatured EspP(46+β) or EspP(46+β)G1066A was incubated with SurA and proteoliposomes containing BamABCDE. Aliquots were placed on ice at various time points, heated to 95°C after the addition of SDS-PAGE buffer, and analyzed by Western blot using an anti-EspP C-terminal peptide antiserum. (B) Quantitation of the data shown in (A). (C) The experiment shown in (A) was repeated with EspP(HA-251+β) or EspP(HA-251+β). (D) Quantitation of the data shown in (C). (E) A second aliquot from the experiment shown in (C) was placed on ice at each time point and treated with PK. After the addition of SDS-PAGE buffer the samples were heated to 95°C and analyzed by Western blot using an anti-HA antiserum.
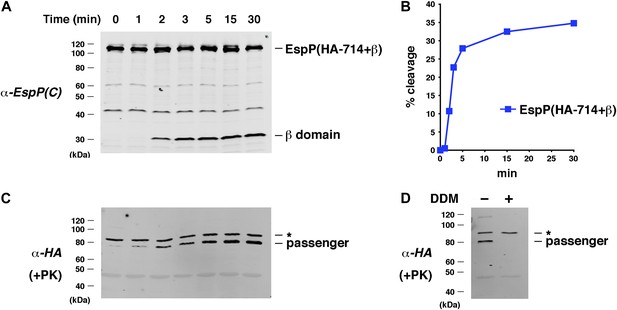
Assembly kinetics of an EspP derivative containing the full-length β helix.
(A) Urea-denatured EspP(HA-714+β) was incubated with SurA and proteoliposomes containing BamABCDE. Aliquots were placed on ice at various time points, heated to 95°C after the addition of SDS-PAGE buffer, and analyzed by Western blot using an anti-EspP C-terminal peptide antiserum. (B) Quantitation of the data shown in (A). (C) A second aliquot from the experiment shown in (A) was placed on ice at each time point and treated with PK. After the addition of SDS-PAGE buffer the samples were heated to 95°C and analyzed by Western blot using an anti-HA antiserum. (D) The experiment described in (A) was repeated. Two equal aliquots were placed on ice after 30 min and DDM was added to one aliquot. The samples were then treated with PK and analyzed by Western blot as described in (C). In (C and D) the asterisk denotes an unidentified background band.
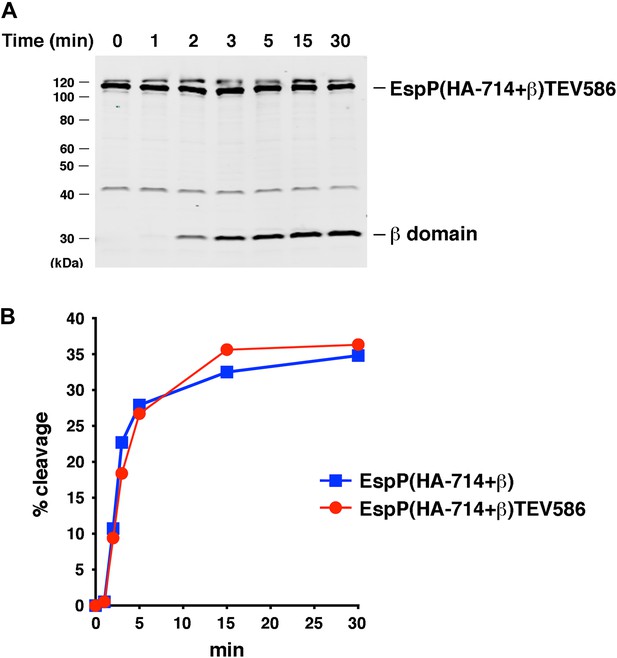
Effect of a linker insertion in the passenger domain on the assembly of EspP(HA-714+β).
(A) Urea-denatured EspP(HA-714+β)TEV586 was incubated with SurA and proteoliposomes containing BamABCDE. Aliquots were placed on ice at various time points, heated to 95°C after the addition of SDS-PAGE buffer, and analyzed by Western blot using an anti-EspP C-terminal peptide antiserum. (B) Quantitation of the data shown in part A (red curve) and Figure 7A (blue curve).
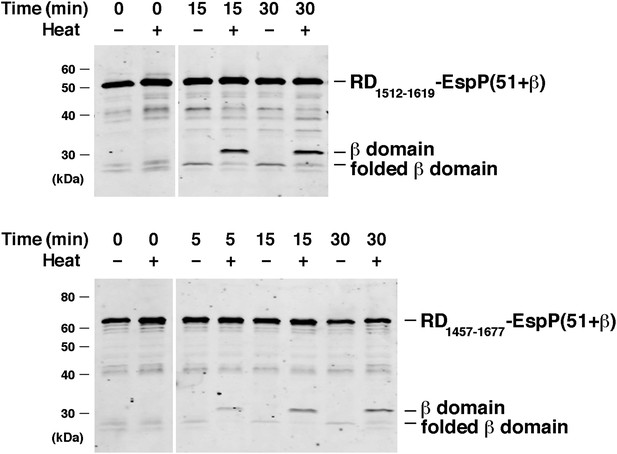
BamABCDE and SurA catalyze the assembly of RD-EspP chimeras.
Urea-denatured RD1512–1619-EspP(51+β) and RD1457–1677-EspP(51+β), which contain 108 and 221 residue fragments of the intrinsically disordered receptor domain (RD) of the B. pertussis RTX toxin fused to the C-terminus of EspP, were incubated with SurA and proteoliposomes containing BamABCDE. Aliquots were placed on ice at various time points, heated to 95°C or maintained at room temperature after the addition of SDS-PAGE buffer, and analyzed by Western blot using an anti-EspP C-terminal peptide antiserum. Chimeric RD-EspP passenger domains have been shown to be secreted efficiently in vivo (Kang’ethe and Bernstein, 2013a).
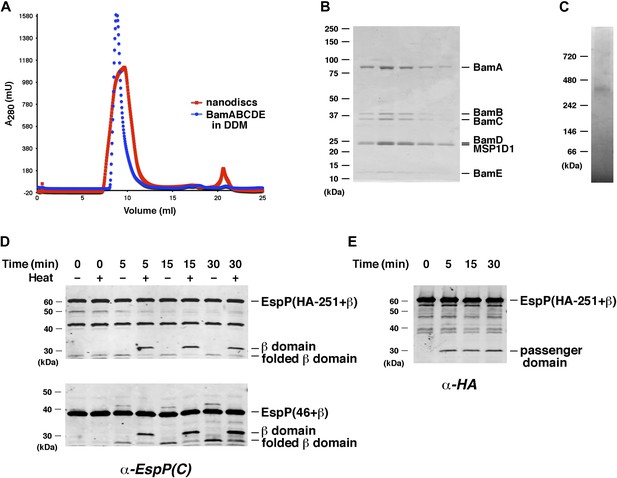
BamABCDE and SurA catalyze the assembly of EspP derivatives into nanodiscs.
(A) Chromatograms of nanodiscs and BamABCDE in DDM on Superdex 75 gel filtration column. (B) SDS-PAGE analysis of the peak fractions in (A). Proteins were visualized by staining the gel with Coomassie Blue. (C) The peak fractions in (B) were pooled and analyzed by Blue Native PAGE. (D) Urea-denatured EspP(HA-251+β) or EspP(46+β) was incubated with SurA and nanodiscs containing BamABCDE. Aliquots were placed on ice at various time points, heated to 95°C or maintained at room temperature after the addition of SDS-PAGE buffer, and analyzed by Western blot using an anti-EspP C-terminal peptide antiserum. The ∼27 kDa polypeptide observed at the 0 min time point on the top gel is an unidentified background band. (E) An additional aliquot from the EspP(HA-251+β) assembly reaction shown in (D) was removed at each time point, heated to 95°C after the addition of SDS-PAGE buffer, and analyzed by Western blot using an anti-HA antiserum.
Additional files
-
Supplementary file 1
Kinetic analysis of EspP assembly in vitro.
- https://doi.org/10.7554/eLife.04234.019
-
Supplementary file 2
Oligonucleotides used in this study.
- https://doi.org/10.7554/eLife.04234.020





