Non-catalytic motor domains enable processive movement and functional diversification of the kinesin-14 Kar3
Figures
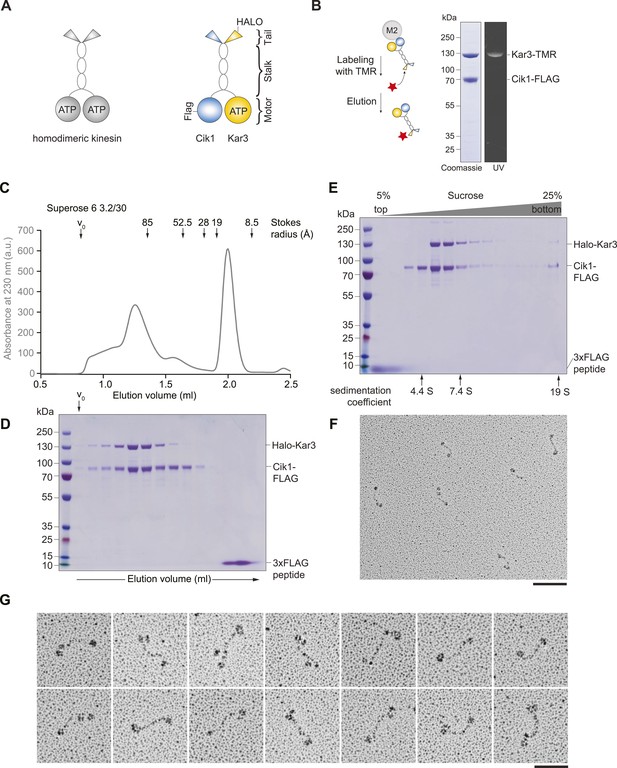
Purification and characterization of Cik1–Kar3 kinesin motors.
(A) Schematic representation of conventional Kinesin-1 in comparison to the kinesin-14 Cik1–Kar3. (B) Purification of recombinant Cik1–Kar3 from yeast extracts. Motors are covalently labeled with Tetramethylrhodamine (TMR) via a HaloTag on the amino-terminus of Kar3. Coomassie-stained SDS-PAGE shows purity of the motor preparation and fluorescent labeling of the Kar3 subunit. (C) Size-exclusion chromatography of Cik1–Kar3-Halo motors on a Superose 6 column. The void volume of the column (V0) and the elution position of standard proteins with their respective stokes radii is indicated. (D) SDS-PAGE analysis of Superose 6 fractions from (C). (E) Sucrose gradient centrifugation of Cik1–Kar3 motors. Consecutive fractions from top to bottom of a 5–25 (wt/vol)% sucrose gradient were analyzed by SDS-PAGE and Coomassie staining. The gradient positions of standard proteins are indicated together with their sedimentation coefficients. (F) Low angle Pt/C rotary shadowing electron microscopy of Cik1–Kar3 motors obtained after size exclusion chromatography. Overview of Cik1–Kar3 motors, scale bar 100 nm. (G) Gallery view of selected Cik1–Kar3 motors, scale bar 50 nm.
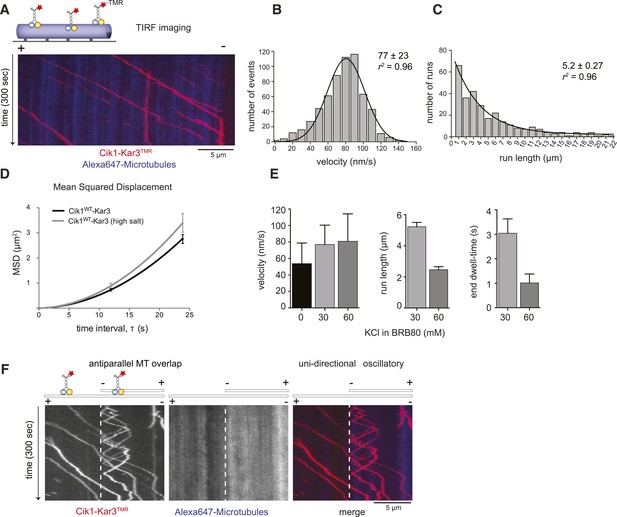
Cik1–Kar3 motors move processively with a single catalytic domain.
(A) Kymograph showing two-color time lapse TIRF microscopy of Cik1–Kar3 (red) moving along taxol-stabilized microtubules (blue). See Video 1 for example of Cik1–Kar3 motility. (B) Histogram of velocities of Cik1–Kar3 molecules moving along taxol-stabilized microtubules (fit with a Gaussian function, black line). The mean velocity is 77 ± 23 nm/s, n = 699. (C) Histogram of run lengths of Cik1–Kar3 molecules moving along taxol-stabilized microtubules, n = 209. (D) Mean-squared displacement analysis of wild-type Cik1–Kar3 at two different salt concentrations in the presence of ATP. (E) Influence of ionic strength on the motile properties of Cik1–Kar3 molecules. Experiments were performed in BRB80-based imaging buffer containing the indicated concentrations of KCl. (F) Behavior of Cik1–Kar3 in microtubule networks. Typical kymograph showing directional movement of Cik1–Kar3 on single microtubules vs repeated directionality switches of individual motors in antiparallel overlaps. The dashed line indicates the beginning of an overlap zone. See Video 2.
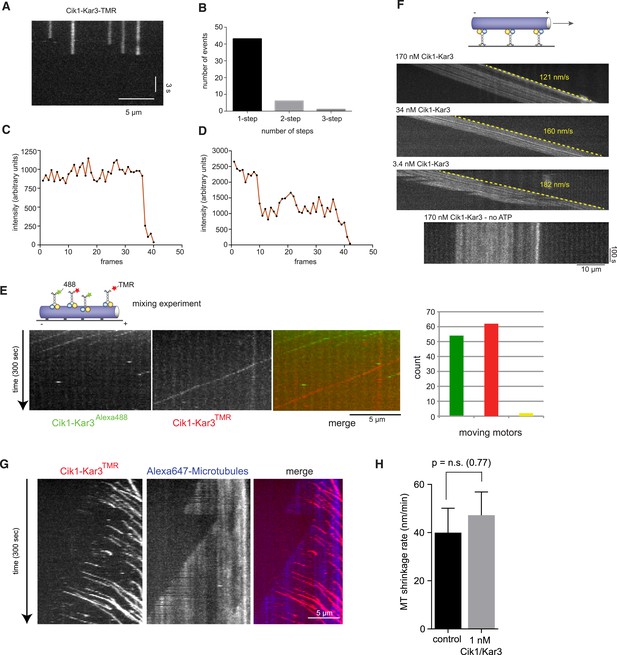
Characterization of Cik1–Kar3 motility.
(A) Photobleaching experiment in the absence of nucleotide. Kymograph representation showing that Cik-Kar3 motors do not exhibit displacement or diffusion in this state. (B) Quantification of photobleaching. (C) Example for single-step photobleaching event. (D) Example for two-step photobleaching event. (E) Mixing experiment combining TMR-labeled Cik1–Kar3 motors with Alexa488 labeled Cik1–Kar3 motors prior to imaging. Kymograph reveals individual traces for red-and green fluorescent motors, indicating that a single Cik1–Kar3 heterodimer is sufficient for movement. (F) Microtubule-gliding experiment with varying concentrations of Cik1–Kar3. In this assay, the motor is immobilized via anti-Halo antibodies to the coverglass and movement of microtubules is recorded by time-lapse TIRF microscopy. Kymographs indicate gliding velocity at different motor concentrations, the lower panel shows that gliding is ATP dependent. (G) Movement of Cik1–Kar3 motors on dynamic microtubules. Dynamic extensions were grown from GMPCPP-stabilized microtubule seeds and imaged together with Cik1–Kar3. Note movement of motors opposite to the directions of microtubule growth, indicating minus-end directed motility of Cik1–Kar3. (H) Quantitative analysis of MT shrinkage rate of taxol-stabilized MTs in the presence of 1 nM Cik1/Kar3 (error bars represent SEM, n = 30).
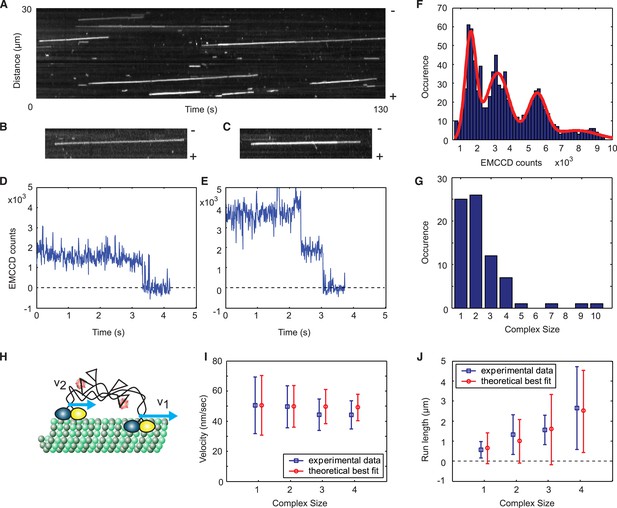
A single Cik1–Kar3 heterodimer is sufficient to form a processive complex.
(A) Kymograph showing movement of Cik1–Kar3. Position along the microtubule is depicted on the vertical axis while time changes along the horizontal axis. (B–C) Kymographs showing binding and movement of single Cik1–Kar3 complexes each composed of 1 (B) or 2 (C) heterodimers. (D–E) Records for background subtracted brightness vs times for complexes in B and C, respectively. (E) Distribution of background subtracted brightness for Cik1–Kar3 complexes. The fit is with four peak Gaussian. (F) Distribution of the initial size of processively moving complexes based on the brightness of a single fluorophore from E. (G) Histogram of moving motor complexes containing the indicated number of Cik1-Kar3 heterodimers. (H) Model of oligomerization of Cik1–Kar3 complexes. Oligomer of two complexes is shown. Blue arrows show velocities of movement of each independent heterodimer. Interconnected tails act as a spring-like linker in between two moving heads. (I, J) Velocity of the movement and run length as a function of size. Blue squares are experimental data, red circles are results of theoretical modeling (for parameters seeFigure 2—figure supplement 3). The bars show average value and standard deviation.
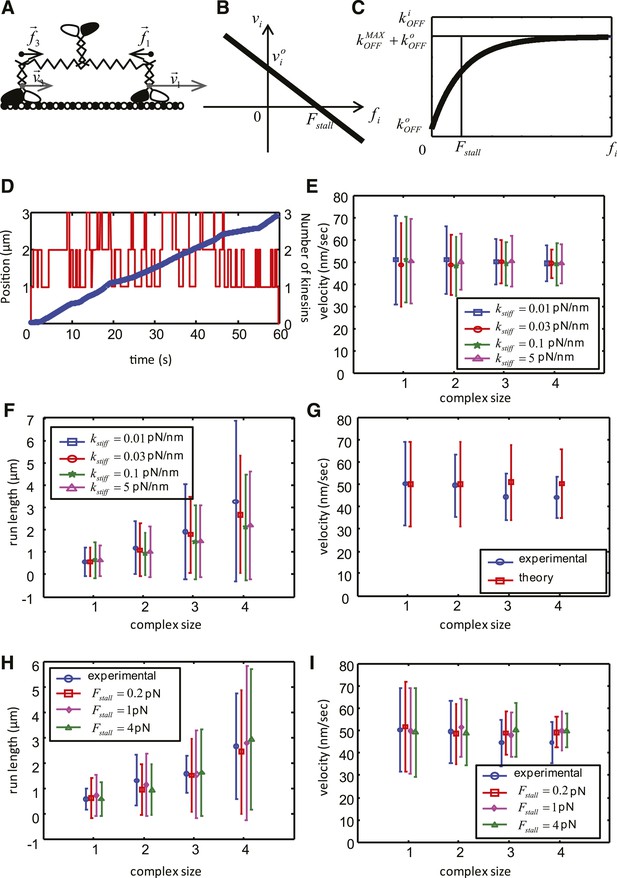
Mathematical model of the cooperative kinesin movement.
(A) Schematics of the kinesin team. In the depicted case, the team consists of three kinesins and two of them are shown as bound to the microtubule and move at the velocities and , respectively. Since the elastic linkage between the dimers stretches generating the forces and . (B) Representation of the force–velocity relation used in the model. (C) Representation of the dependence of the unbinding rate on the applied force. (D) Example of a simulation for a complex size with three dimers. The blue graph is the position of the center of mass of the complex as a function of time (left axis), red graph shows the number of dimers attached to the microtubule at each time point. (right axis). (E–F) Velocity, run length and their standard deviations as a function of the complex size for different values of . Other parameters of the simulation are: V = 50 nm/s, = 20 nm/s; = 1 pN; = 0.55 s−1; = 0.08 s−1; = 4 s−1. (G) Comparison between experimental data (blue) and best fit (see values of objective function in Table 5) of the simplified theoretical model without force dependence (red). (H–I) Velocity, run length and their standard deviations as a function of the complex size for best parameters of the complete model. Data are shown for red— = 0.2 pN; = 0.57 s−1; = 3.7 s−1; magenta—= 1 pN; = 0.55 s−1; = 4 s−1; green— = 4 pN; = 0.49 s−1; = 5.3 s−1; Other parameters for all simulations: V = 50 nm/s, = 20 nm/s; = 0.08 s−1; = 0.03 pN/nm.
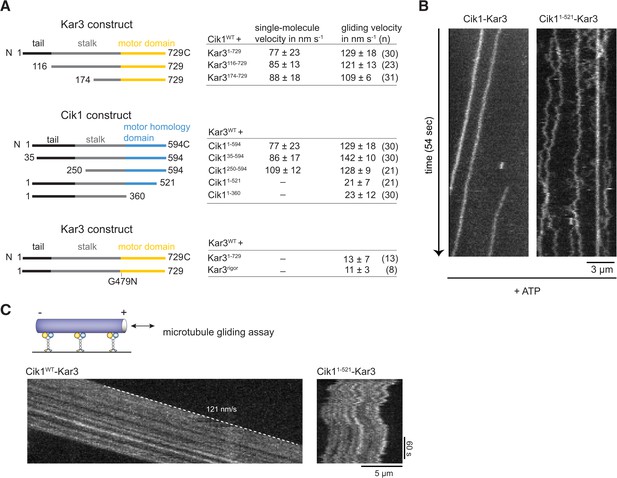
Molecular requirements for processivity and identification of a translocation-deficient Cik1 mutant.
(A) Schematic showing analyzed Cik1 and Kar3 truncation constructs with the corresponding results from TIRF assays and multi-motor gliding assays. All constructs contained the Halo-tag at the aminoterminus of Kar3 for fluorescent labeling with TMR. (B) Kymographs of TMR-labeled Kar3 complexes containing either full-length Cik1 (aa 1–594) or the carboxyterminal truncation mutant Cik11–521. See Videos 4 and 5. (C) Kymograph of microtubule-gliding by full-length Cik1–Kar3 and the Cik11–521-Kar3 mutant. Note the impairment of directional translocation by the Cik11–521–Kar3 mutant.
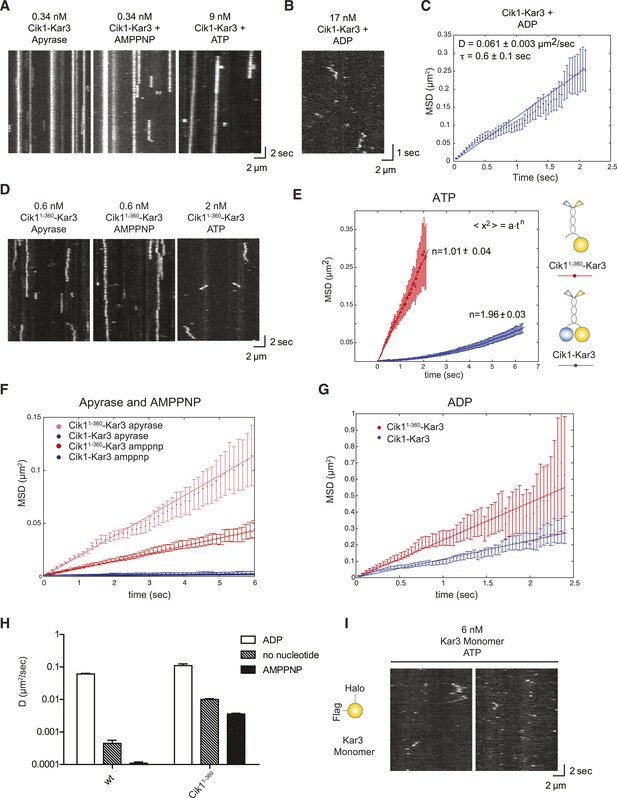
Single-molecule analysis of Kar3 motors in different nucleotide states.
(A) Kymographs showing single molecule TIRF microscopy of Cik1–Kar3 motors in different nucleotide states. Videos were taken with high temporal resolution (35 frames per second). Note the different motor concentrations and the directional displacement of Cik1–Kar3 molecules in the presence of ATP. (B) Diffusive movement of individual Cik1–Kar3 molecules in the presence of ADP. (C) MSD analysis of the diffusive movement of Cik1–Kar3 motors in the presence of ADP. Molecules with lifetimes between 0.5 and 5 s were analyzed. (D) Typical kymographs of Cik11–360–Kar3 motors lacking the non-catalytic head domain in no-nucleotide (Apyrase), AMPPNP and ATP states. Note the diffusive interactions of the motor with the microtubule in comparison to 3A. (E) Mean-squared displacement (MSD) analysis of Cik1–Kar3 and Cik11–360–Kar3 motors in the presence of ATP. Data points were fitted to the formula <x2> = a·tn. n = 1 for Cik11–360–Kar3 indicates random diffusion without bias and constraints, while n = 2 for the wild-type motor indicates directional, processive movement. (F) Quantitative comparison of microtubule interactions of wild-type vs Cik11–360–Kar3 motors in no nucleotide and AMPPNP states by mean squared displacement analysis. (G) MSD analysis of wild-type vs Cik11–360–Kar3 motors in the ADP state. (H) Summary of the diffusion coefficients obtained for wild-type vs Cik11–360-Kar3 motors in three different nucleotide states. Note the logarithmic scale on the y-axis. (I) Typical kymographs of a monomeric Kar3 head construct (residues 353–729 fused to an N-terminal Halo tag) in the presence of ATP.
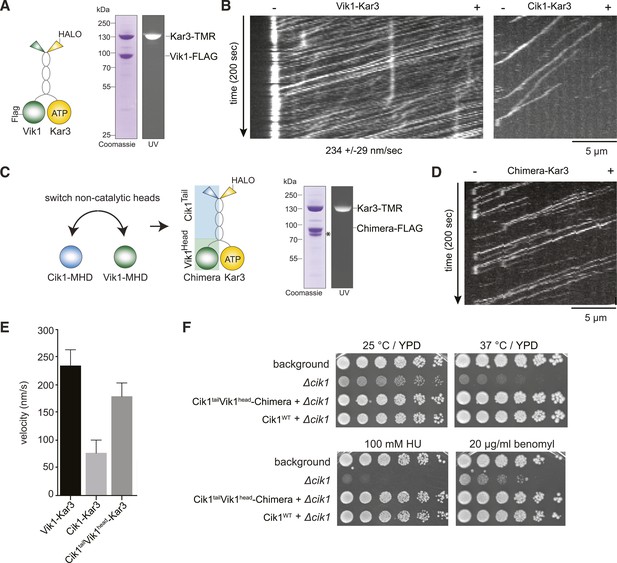
The non-catalytic head domain determines the velocity of the Kar3 motor.
(A) Schematic representation of the kinesin-14 Vik1–Kar3 and purification of recombinant Vik1–Kar3 from yeast extracts. Motors are covalently labeled with Tetramethylrhodamine (TMR) via a HaloTag on the amino-terminus of Kar3. Coomassie-stained SDS-PAGE shows homogeneity of the motor preparation and fluorescent labeling of the Kar3 subunit. (B) Kymographs of single color time lapse TIRF microscopy highlighting the velocity difference between Vik1–Kar3 and Cik1–Kar3 when moving along taxol-stabilized microtubules. (C) Construction and purification of a chimeric motor in which the non-catalytic heads were switched. An asterisk denotes a proteolytic degradation product. (D) Kymograph of typical single molecule runs by the chimeric Cik1Tail–Vik1Head motor. (E) Comparison of velocities of the different Kar3 constructs. (F) Serial dilution growth assay of the indicated yeast strains at different temperatures and conditions. Note that the integration of the chimeric Cik1Tail–Vik1Head fusion protein fully rescues the phenotypes of a Cik1 deletion.
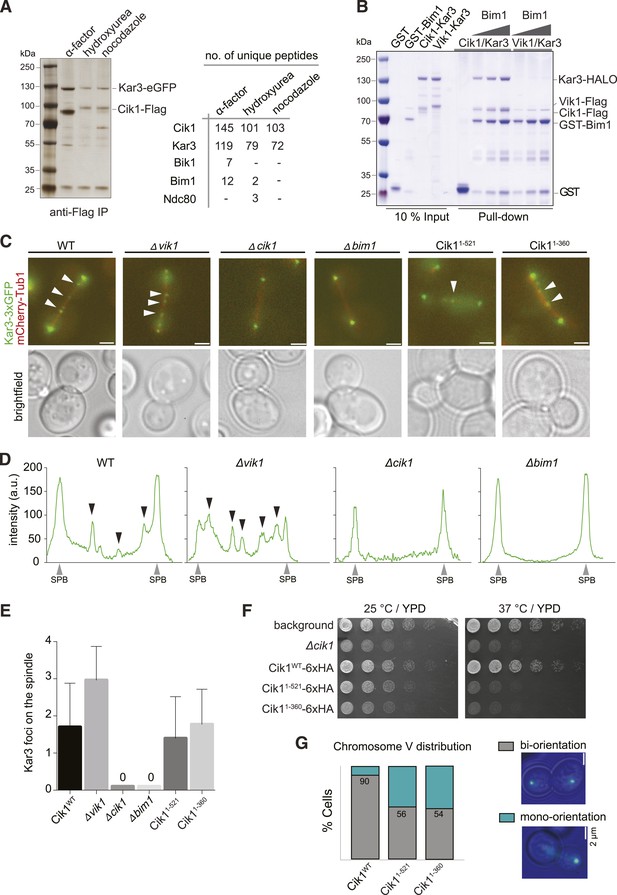
Cik1 specifies differential interactions of Kar3 motors to determine their subcellular localization.
(A) Affinity purification of the Cik1–Kar3 complex from different cell cycle states and identification of interaction partners by mass spectrometry. (B) Pull-down assay with GST-Bim1 and Cik1- or Vik1-Kar3. Only Cik1-Kar3 interacts with Bim1 in a dose-dependent manner. (C) Localization of Kar3 in different deletion mutants in yeast investigated by live cell microscopy. (D) Panel shows line scans of Kar3–GFP intensity along the spindle axis. Arrowheads point to Kar3 foci along the spindle. Scale bar: 3 μm. Both Cik1 and Bim1 are required for the localization to the anaphase spindle. (E) Quantification of Kar3 foci on anaphase spindles in the indicated yeast strains. Kar3 spindle signals were never detected in a bim1Δ or cik1Δ strain. (F) The Cik11–521 and Cik11–360 mutants elicit a temperature-sensitive phenotype in vivo. Growth assay was performed with serial dilutions of the indicated yeast strains at different temperature on rich medium (YPD). (G) Quantification of chromosome segregation in the indicated yeast strains at 25°C, n = 100 for all strains analyzed. Right panel shows representative fluorescent micrographs with segregation of fluorescently labeled Chromosome V.
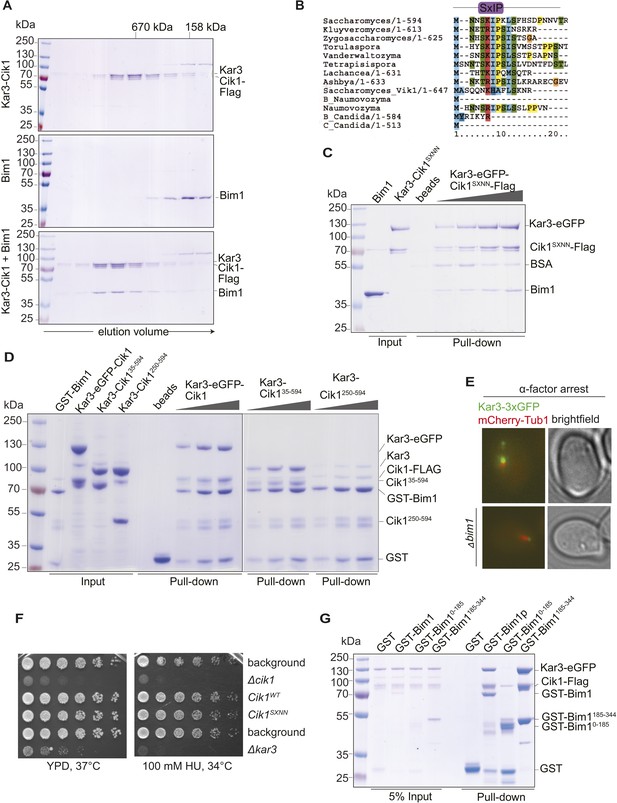
Biochemical and genetic characterization of the Cik1-Kar3-Bim1 interaction.
(A) Analytical size exclusion chromatography of Cik1–Kar3 and Bim1 individually or in combination. Shift to earlier elution position indicates complex formation. (B) Sequence alignment of the amino-termini of selected Cik1 and Vik1 proteins from diverse yeasts. Note the presence of an SxIP motif (EB1 interaction motif) in Cik1 proteins that is absent from Vik1. (C) Mutation of the SxIP motif in Cik1 does not impair binding of the Cik1–Kar3 complex to Bim1 in GST-pull down assays. (D) The more extended amino-terminus of Cik1 is required for Bim1 binding. GST-Bim1 pull-down assays with indicated Cik1–Kar3 truncation mutants. Elimination of the first 250 amino acids in Cik1 impairs binding of the motor complex to Bim1. (E) Localization of Kar3 to the tips of shmoo-directed microtubules in alpha-factor arrested cells depends on Bim1. Live cell microscopy of Kar3-3xGFP (green) in either wild-type (upper panel) or bim1-deletion mutant (lower panel). Note the lack of microtubule plus-end localization of Kar3 in the bim1 deletion mutant. (F) Mutation of the conserved SxIP motif in Cik1 does not cause a growth phenotype in vivo. Spot assays were carried out with the indicated strains either on YPD plates or on YPD plates supplemented with 100 mM Hydroxyurea. (G) Binding of Cik1–Kar3 to Bim1 occurs via the cargo-binding domain. GST-pull down assays with full-length Bim1 or versions constituting the microtubule (Lampert et al., 2013) in the absence of the motor.
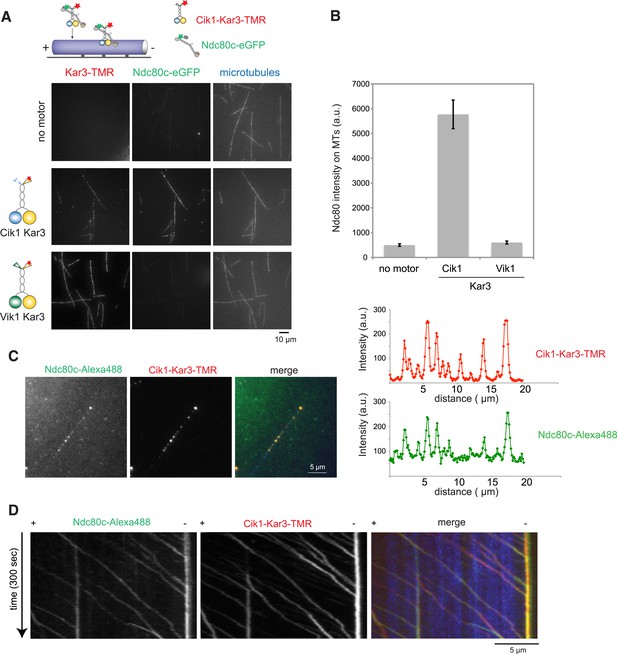
Processive transport of the Ndc80 kinetochore complex by Cik1–Kar3 motors in vitro.
(A) Cik1–Kar3, but not Vik1–Kar3 recruits the Ndc80 kinetochore complex to taxol-stabilized microtubules in vitro. The experiment was performed with TMR-labeled Kar3 motors and fluorescently labeled recombinant Ndc80 complex in the absence of nucleotide. (B) Quantification of the recruitment experiment by analyzing fluorescence intensity of microtubule-bound Ndc80-eGFP complex in the presence of different motor constructs. Error bars denote standard error of the mean (s.e.m.) (C) Co-localization of the Ndc80 kinetochore complex and Cik1–Kar3 motors to distinct signals on the microtubule lattice. Right panel shows line scans of fluorescent intensity of Kar3 motors and Ndc80 complex along the length of a microtubule. (D) Kymograph showing triple-color time-lapse TIRF microscopy demonstrating that in the presence of ATP, Ndc80 complexes are processively transported towards microtubule minus ends by Cik1–Kar3 motors.
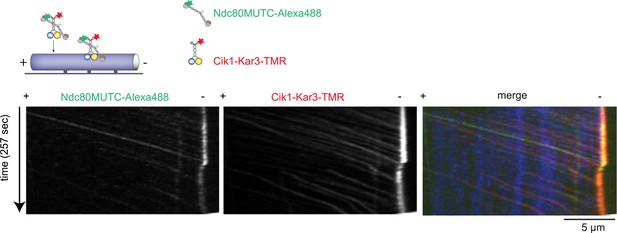
Cik1–Kar3 transports a mutant Ndc80 kinetochore complex that is unable to bind MTs.
Schematic and kymographs of an effective recruitment and translocation of the microtubule-binding defective calponin-homology domain mutant K122E K204E Ndc80 complex by the motor Cik1–Kar3. The experiment was performed with TMR-labeled Kar3 motors, Alexa647-taxol stabilized MTs, and fluorescently labeled recombinant Ndc80 complex in the presence of ATP.
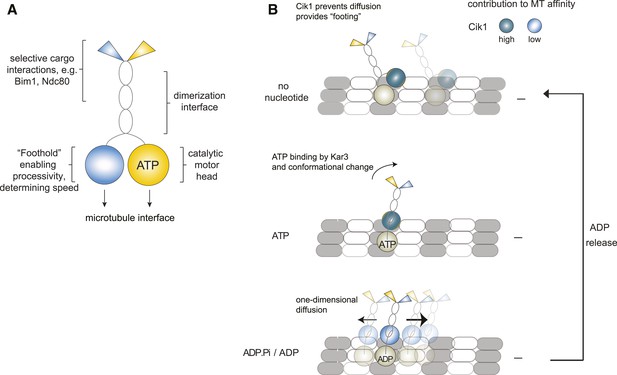
Model for Kar3 motility and function.
(A) Functional contributions of the different domains of heterodimeric Kar3 motors analyzed in this study. (B) Model for Cik1–Kar3 motility depending on a non-catalytic head domain. Upon ATP uptake a conformational change occurs with the stalk rotating towards the minus end. A key role for the non-catalytic domain is to prevent diffusion in this state but allow one-dimensional diffusion in the subsequent ADP state with a bias towards the minus end.
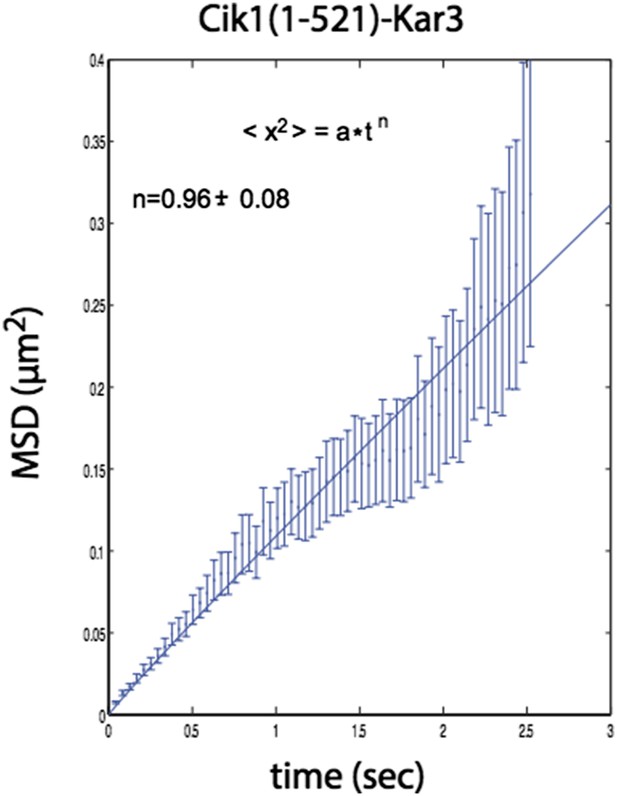
n=1 for the fit indicates that the movement of this mutant also does not contain a directional component.
Videos
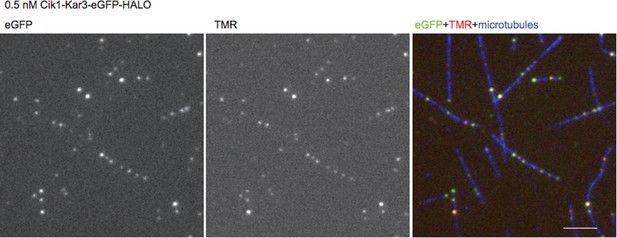
Time-lapse two-color TIRF microscopy of Cik-Kar3-TMR motors (red) moving on taxol stabilized microtubules (blue). 100 frames were taken every 3 s. The video is played at 20 frames/s, scale bar: 5 μm. The video corresponds to Figure 2A.
https://doi.org/10.7554/eLife.04489.008Behavior of Cik1–Kar3 in microtubule overlap zones. Two-color time-lapse TIRF video of Cik1–Kar3-TMR (red) moving on taxol-stabilized microtubules (blue). Note back and forth movement of individual Cik1–Kar3 motors in microtubule overlap zones. 100 frames were taken every 3 s, the video is played at 20 frames/s, scale bar 5 μm. The video corresponds to Figure 2F.
https://doi.org/10.7554/eLife.04489.009Mixing experiment to demonstrate processivity of individual Cik1–Kar3 heterodimers. Cik1–Kar3-TMR motors (red) were mixed with Cik1–Kar3-Alexa488 motors (green) and imaged by multi-color TIRF microscopy on taxol-stabilized microtubules. Red and green motors are seen moving in opposite directions because of closely spaced microtubules with opposite orientation. Frames were taken every 3 s for 100 frames, the video is played at 20 frames/s. Scale bar corresponds to 5 μm. The video corresponds to Figure 2—figure supplement 1E.
https://doi.org/10.7554/eLife.04489.010Wild-type Cik1–Kar3-TMR in the presence of 5 mM ATP moving on taxol-stabilized microtubules imaged at higher frame rate in single color with TIRF microscopy. Time between frames is 273 ms, the frames are numbered in the upper left corner, total length of the video is 54 s. The video is played at 20 frames/s. Scale bar is 3 μm. The video corresponds to Figure 3B.
https://doi.org/10.7554/eLife.04489.012Cik1–Kar3-TMR with truncation of the non-catalytic Cik1 globular domain (Cik1 1–521) interacting with taxol-stabilized microtubules in the presence of 5 mM ATP. Frame rate and imaging conditions as in Video 5. Note non-directional movement of Cik11–521-Kar3. Scale bar 3 μm. The video corresponds to Figure 3B.
https://doi.org/10.7554/eLife.04489.013Vik1–Kar3 motors display processive movement and pronounced minus-end dwelling. Two-color time lapse TIRF microscopy of Vik1-Kar3-TMR (red) on taxol-stabilized microtubules (blue). Note strong accumulation of motors at minus-ends at the end of the video. 100 frames were taken every 3 s, video is played at 20 frames/s, scale bar is 5 μm. The video corresponds to Figure 5B.
https://doi.org/10.7554/eLife.04489.016Transport of the Ndc80 kinetochore complex by Cik1–Kar3 motors along taxol-stabilized microtubules. Triple-color TIRF video showing Ndc80-Alexa488 complex (green), Cik1–Kar3-TMR motors (red), and Alexa647-Taxol stabilized microtubules (blue) in the presence of 5 mM ATP. Frames were taken every 3 s. Note co-transport of Ndc80 complexes and Cik1–Kar3 complexes towards the end of the microtubule. Video is played at 20 frames/s, scale bar is 5 μm. The video corresponds to Figure 7D.
https://doi.org/10.7554/eLife.04489.021Additional files
-
Supplementary file 1
This file contains a detailed description of the mathematical model for cooperative Cik1–Kar3 kinesin movement.
- https://doi.org/10.7554/eLife.04489.023
-
Supplementary file 2
This file contains tables listing the yeast strains and plasmids used in this study.
- https://doi.org/10.7554/eLife.04489.024