Spectrin binding motifs regulate Scribble cortical dynamics and polarity function
Figures
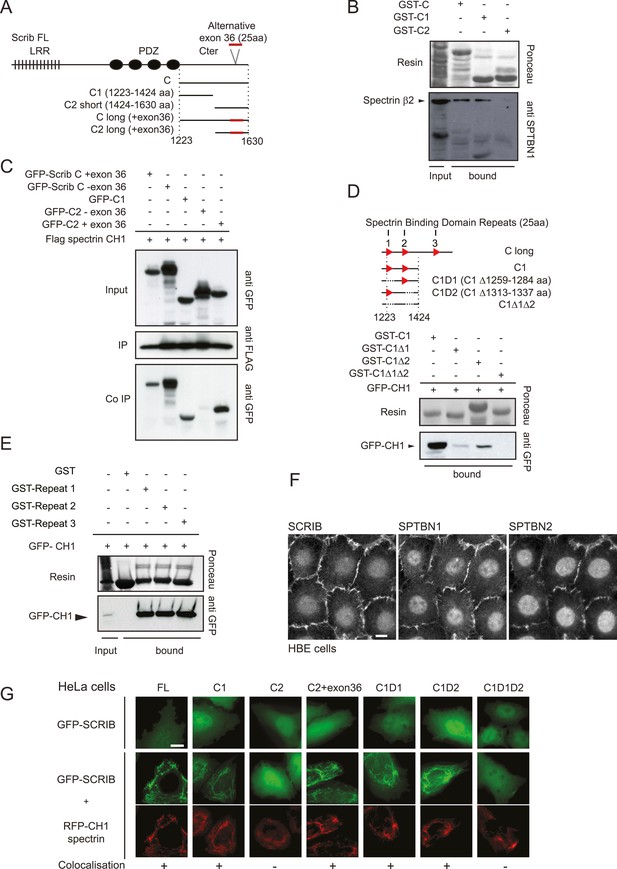
Dissection of the SCRIB β-spectrin interaction.
(A) Schematic representation of SCRIB and different C-terminal constructs used in this study. (B) GST-C, GST-C1 and GST-C2 pull-down on astrocytes cell extract. Samples were analyzed by ponceau staining and immunoblotting using the indicated antibody. (C) Immunoprecipitation was performed with anti Flag antibody using HEK293 cell lysates co-expressing Flag CH1 spectrin domain with indicated GFP-SCRIB constructs. Samples were analyzed by immunoblotting using the indicated antibodies. (D) Schematic representation of the C1 SCRIB constructs with internal deletions of putative spectrin binding motif sequences. GST-C1, GST-Δ1, GST-Δ2, GST-Δ1Δ2 pull-down assay on HEK293 cell lysates expressing GFP-CH1 spectrin domain. Samples were analyzed by immunoblotting using anti GFP. (E) GST and GST-SCRIB repeat 1, 2 and 3 pull down assay on HEK293 cell lysates expressing GFP-CH1 spectrin domain. Samples were analyzed by ponceau staining and immunoblotting using the indicated antibody. (F) Immunofluorescence images of 16HBE monolayers fixed and stained for SCRIB, SPTBN1 (spectrin β2) and SPTBN2 (spectrin β3). (G) HeLa cells were singly transfected with indicated GFP-SCRIB constructs (upper panel) or in combination with spectrin RFP-CH1 domain (bottom panels). Images are representative of at least three independent experiments. Bars, 10 µm.
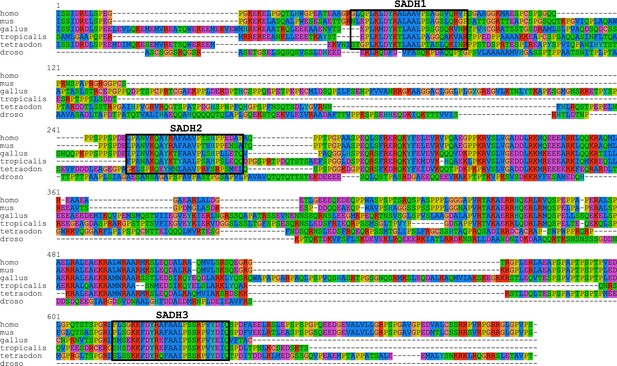
SCRIB C-terminal sequence alignment.
Sequence alignment of SCRIB C-terminal domain of indicated different species. Boxes indicate the emplacement of the SADH motifs. Note that no sequences fitting the SADH consensus can be detected in Drosophila Scribble C-terminal domain sequence.
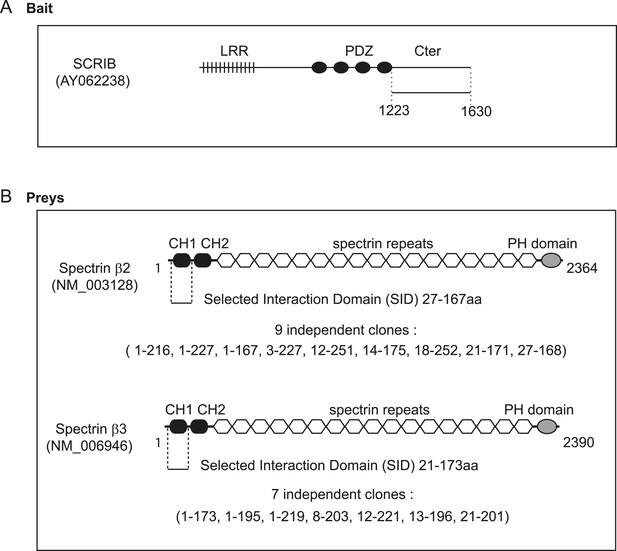
Schematic representation of SCRIB, β2-spectrin and β3 spectrin proteins.
(A) The SCRIB C-terminal sequence encompassing the 1223–1630 region was used as a bait in the two hybrid screen. (B) Minimal β2-spectrin and β3 spectrin sequences recovered in the two hybrid screen are indicated (SID) as well as the exact start and stop of each individual selected clones.
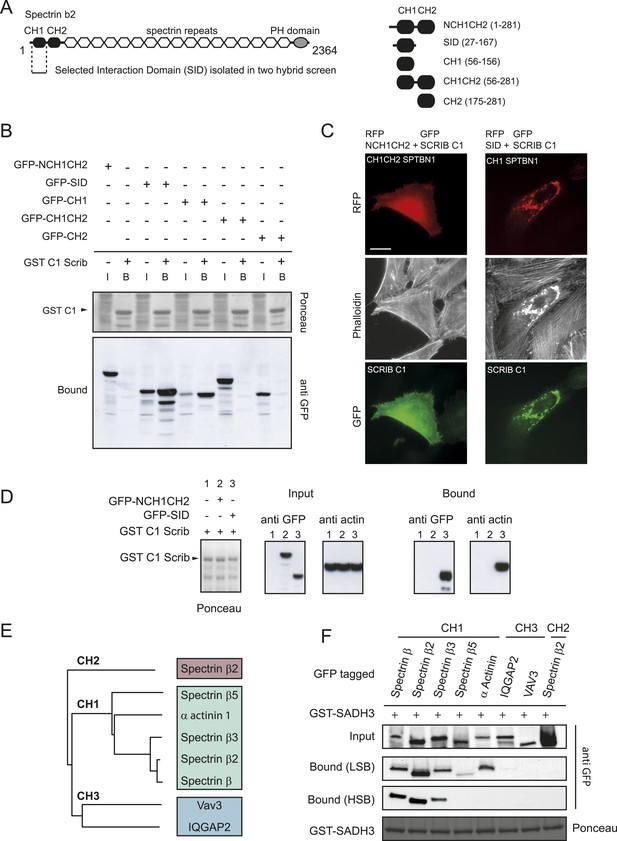
SCRIB spectrin binding motifs bind to CH1 domains of the β spectrin family.
(A) Schematic representation of full length human β2 spectrin protein (left) and different β2 spectrin deletion constructs used in the study (right). (B) GST pull down using a GST-SCRIB C1 resin from HEK293 cell lysates expressing indicated GFP-tagged β2 spectrin CH domains. Samples were analyzed by ponceau staining and immunoblotting using anti GFP. (C) Fluorescence images showing HeLa cells expressing GFP-SCRIB C1 construct together with RFP-NCH1CH2 or SID β2 spectrin domains and stained with phalloidin. (D) GST-SCCRIB C1 pull down from HEK293 cell lysates expressing indicated GFP tagged β2 spectrin CH domains. Samples were analyzed by ponceau staining and immunoblotting using anti GFP. (E) Phylogenetic analysis of the Calponin Homology (CH) domains type1, 2 and 3 used in this study. (F) GST pull down from HEK293 cell lysates expressing the indicated CH domains using a GST-SCRIB SADH3 resin. Pull down was done in low or high stringency conditions (LSB: Low Salt Buffer, HSB: High Salt Buffer). Ponceau staining indicates the relative amount of GST tagged proteins bound to the resin. Samples were analyzed by ponceau staining and immunoblotting using anti GFP. Bars, 10 µm.
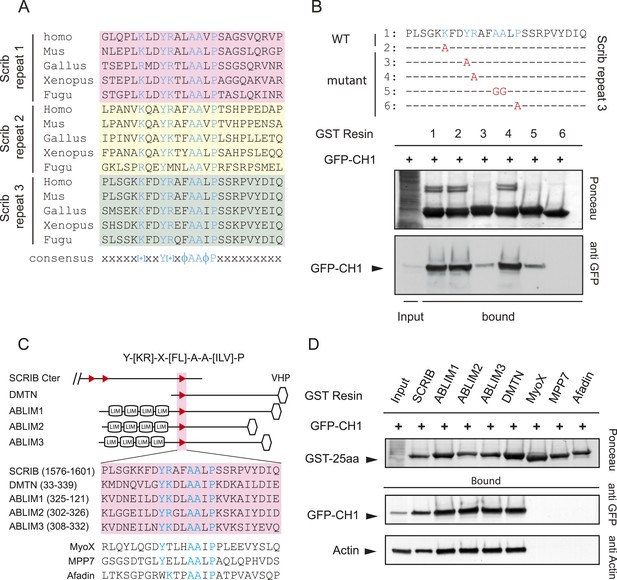
The SADH motif (SCRIB, ABLIM, dematin homology motif).
(A) Alignment of vertebrate SCRIB spectrin binding repeats 1 (pink), 2 (yellow) and 3 (green). Conserved residues are shown in blue. [+] indicates positively charged amino acids and Φ represents hydrophobic residues. (B) Alanine or glycine substitutions (in red) performed on SCRIB repeat 3 conserved residues. WT and mutants GST-repeat 3 pull down assay on HEK293 cell lysates expressing spectrin GFP-CH1 domain. Samples were analyzed by ponceau staining and immunoblotting using the indicated antibody. (C) Schematic representation and sequence of DMTN and ABLIM1, 2 and 3 proteins displaying strong homology with the SCRIB putative spectrin binding motif Y-[KR]-X-[FL]-A-A-[ILV]-P (boxed in pink). Position of the motif in the protein is indicated in brackets in aa. MyoX, MPP7 and Afadin sequences with noncanonical motifs consensus are shown on the bottom. VHP (Villin Hedapiece domain). (D) Pull down assay with GST resin fused to 25aa motif from indicated proteins performed in HEK293 cell lysates expressing spectrin GFP-CH1 domain. Samples were analyzed by ponceau staining and immunoblotting using the indicated antibody.

SADH domains in vertebrates.
(A) Schematic representation of SCRIB gene with spectrin binding motifs (up). Table displaying the distribution of SADH domains in SCRIB vertebrate protein sequences. SADH motifs could not be identified in drosophila and C. elegans Scribble/LET-413 ortholog genes. (B) Schematic representation of SADH motifs in DMTN and in ABLIM123 genes (up) and conservation of SADH motif sequence in vertebrates.
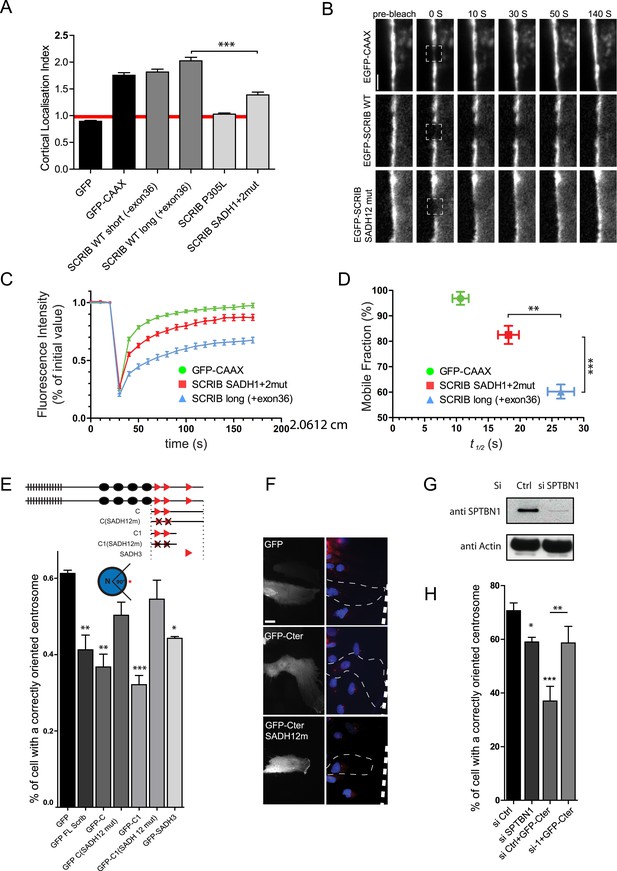
SADH motif influence SCRIB membrane stability and is required for SCRIB polarity function.
(A) 16HBE cells were transiently nucleofected with the indicated GFP constructs and analyzed by live confocal microscopy to calculate their cortical localization index. (n = 50 for each conditions). (B) FRAP experiment on 16HBE cells nucleofected with the indicated GFP constructs. High magnification images of adherens junction before and at the indicated time points after photobleaching. (C) Quantitative analysis of FRAP from experiments similar to those shown in B (n = 30 for each conditions). (D) The mobile fraction and t1/2 of recovery for each protein were calculated from the recovery curves in C. (E) Centrosome reorientation in migrating astrocytes expressing the indicated DNA constructs (up). Results shown are means SEM of 3–4 independent experiments, with a total of at least of 150 cells. (F) Representative images of astrocytes expressing the indicated constructs and stained with anti-pericentrin (centrosome, red) and Hoechst (nucleus, blue). (G) Primary astrocytes were nucleofected with control (Ctrl) or β2 spectrin (si SPTBN1) siRNA and incubated for 72 hr. Protein levels were analyzed by western blots using anti-SPTBN1 antibody and anti-actin. (H) Centrosome reorientation assay in migrating astrocytes nucleofected with control or β2 spectrin (SPTBN1) siRNA and microinjected with GFP SCRIB Cter construct. Results shown are means ± SEM of three independent experiments, with a total of at least 100 cells. ***p < 0.001; **p < 0.01; *p < 0.05. Bars, 10 µm.

Cortical localization of SCRIB SADH mutant.
(A) Representative images of 16HBE cells nucleofected with the indicated GFP constructs acquired live and used for the CLI analysis. (B) Schematic representation of the process used for CLI calculation.

SADH domains are implicated in SCRIB-mediated control of Cdc42 localization.
(A) Cherry-Cdc42 recruitment at the leading edge of migrating astrocytes cells expressing the indicated constructs. Results as shown are means ± SEM of three independent experiments, with a total of at least 200 cells. Representative images are shown on the right. White dotted lines show scratch positions. The scale bar represents 10 μm.
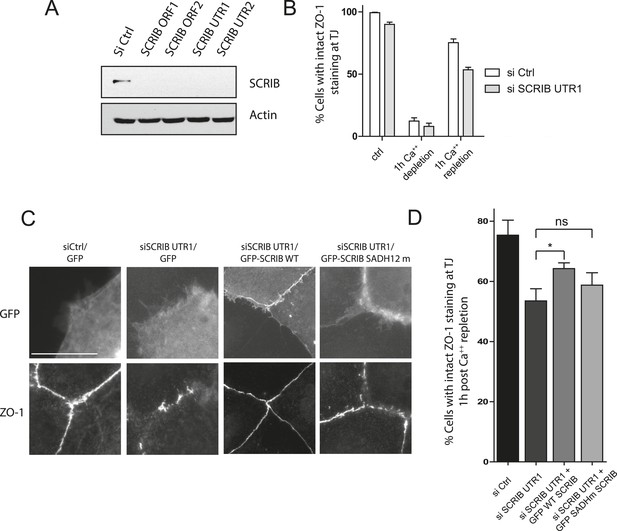
SCRIB SADH domain are required for tight junction assembly.
(A) WB analysis of SCRIB and actin expression in Caco-2 cells, 3 days after nucleofection with control siRNA (Ctrl) or siRNAs directed against SCRIB ORF (ORF1 and ORF2) or 3′ UTR (UTR1 and 2). (B) Percentage of Caco-2 cells nucleofected with control or SCRIB UTR1 siRNA displaying intact of ZO-1 labeling at the cell–cell contacts. (C) Representative example of Caco-2 cells displaying intact or short and disconnected areas of ZO-1 labeling at the cell–cell contacts in the indicated conditions. (D) Caco-2 cells nucleofected with indicated siRNA or construct were scored 1 hr after calcium switch for their tight junction integrity using ZO-1 staining. Results shown are means ± SEM of three independent experiments, with a total of at least 100 cells. The scale bar represents 10 μm.
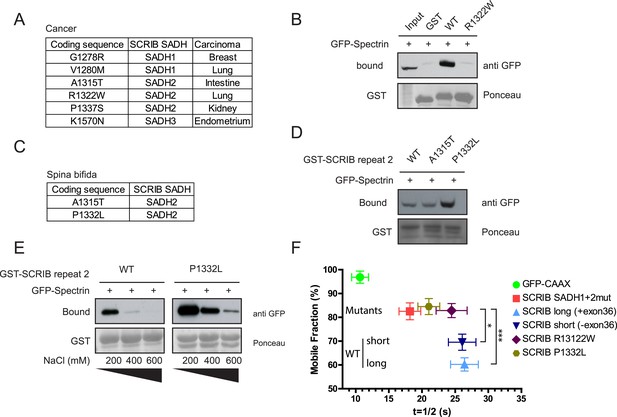
SADH motifs mutations in human pathology.
(A) Table of identified somatic SCRIB SADH motifs mutations in human cancers. Data sourced from COSMIC (http://cancer.sanger.ac.uk/cosmic). (B) GST-SCRIB SADH2 WT and SADH2 R1322W pull down assay on HEK293 cell lysates expressing spectrin GFP-CH1 domain. Ponceau staining indicates the relative amount of GST tagged proteins bound to the resin. Samples were analyzed by immunoblotting using anti GFP. (C) Table of identified germinal SCRIB SADH motifs mutations in spina bifida. (D) GST-SCRIB SADH2 WT, A1315T and P1332L pull down assay on HEK293 cell lysates expressing spectrin GFP-CH1 domain. Ponceau staining indicates the relative amount of GST tagged proteins bound to the resin. Samples were analyzed by immunoblotting using anti GFP. (E) GST-SCRIB SADH2 WT and P1332L pull down assays similar to D performed in indicated salt stringency. (F) Quantitative analysis of FRAP experiments in 16HBE adherens junction expressing the indicated GFP constructs (n = 30 for each conditions). The mobile fraction and t1/2 of recovery for R1322W and P1332L proteins were calculated from the recovery curves in Figure 5—figure supplement 1.
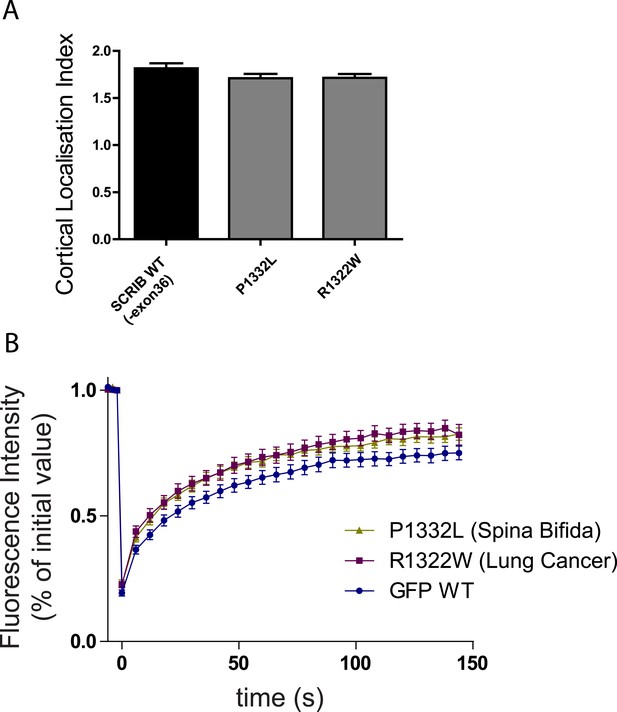
(A) 16HBE cells were transiently nucleofected with the indicated GFP constructs and analyzed by live confocal microscopy to calculate their cortical localization index.
(n = 50 for each conditions). (B) Quantitative analysis of FRAP experiment on 16HBE cells nucleofected with the indicated GFP constructs (n = 30 for each conditions).





