The unfolded protein response is required for dendrite morphogenesis
Figures
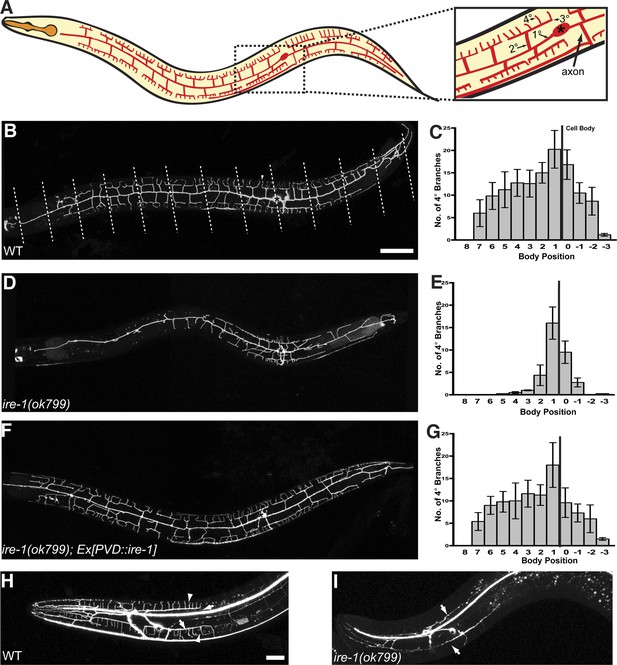
ire-1 is required for PVD dendritic morphogenesis.
(A) Cartoon showing the PVD dendritic arbor. The dash-boxed region is magnified to show the PVD soma (asterisk), axon, primary dendrite (1°), secondary dendrite (2°), tertiary dendrite (3°) and quaternary dendrite (4°). (B) Representative wild type (WT) dendritic morphology of PVD neuron expressing membrane associated mCherry (wyIs581). Starting from the cell body, the anterior and the posterior sections of the primary dendrite are divided into 8 and 4 equal segments, respectively, indicated by dashed lines. Anterior, left; dorsal, top. Asterisk, cell body; arrowhead, quaternary (4°) dendrite. Scale bars, 50 μm. (C) Quantification of the number of quaternary (4°) branches in each segment in WT. The position of cell body is indicated by the black line. Error bars show mean ± s.e.m., n = 10. (D to G) Defective PVD dendritic morphogenesis in ire-1 (ok799) mutants (D and E) is rescued by expressing ire-1 cell-autonomously (F and G). (H and I) Representative dendritic morphology of FLP neurons labeled by cytoplasmic GFP in wild-type (H) and ire-1 (ok799) mutants (I). Asterisks, cell bodies; arrows, secondary branches; arrowheads, tertiary branches. Scale bar, 20 μm.

Schematic diagrams of IRE-1 dependent UPR pathway and of C. elegans IRE-1 protein showing three mutations.
(A) Cartoon showing the IRE-1 dependent UPR pathway. Two missense mutations in wy762 and wy782 are shown by asterisks (B) IRE-1 contains a luminal domain (blue), trans-membrane (TM) domain (black line), a linker (yellow), a S/T kinase domain (red) and an endoribonuclease domain (green). Two missense mutations (wy762 and wy782) are shown by asterisks and the deleted fragment in the null mutation ok799 is illustrated by the gray bar.
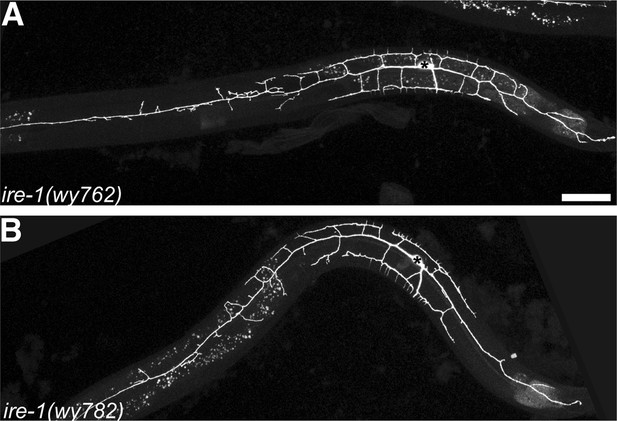
The ire-1 mutants showing PVD dendritic morphogenesis defect.
Representative dendritic morphology of PVD neuron expressing membrane associated GFP (wyIs378) in two ire-1 alleles wy762 (A) and wy782 (B) isolated from forward genetic screen. Asterisks, cell bodies; scale bar, 50 μm.
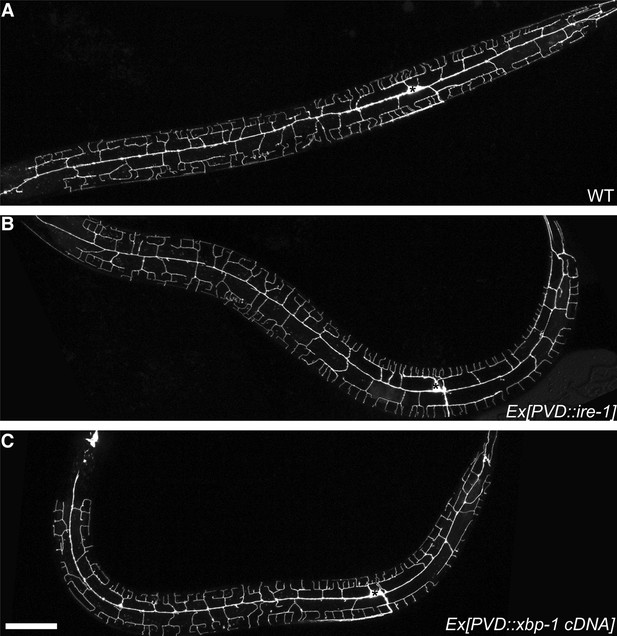
Compared with wild type animal (A), expressing ire-1 cDNA (B) or spliced xbp-1 cDNA (C) did not cause overbranching of PVD in wild type animals.
Asterisks, cell bodies. Scale bar, 50 μm.
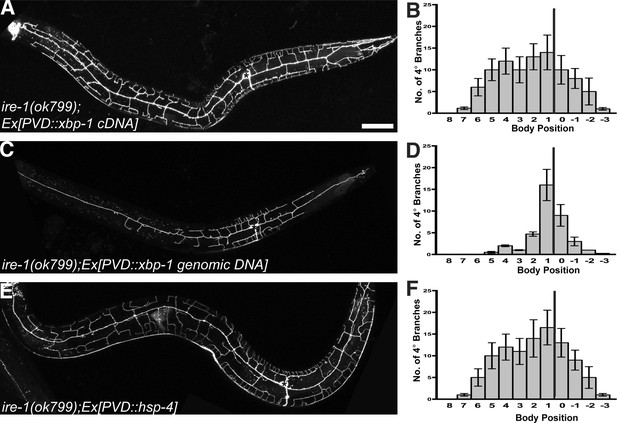
The UPR is required for PVD dendritic morphogenesis.
Expressing xbp-1 cDNA in PVD rescues the defective dendritic morphogenesis in ire-1 mutants (A and B) while expressing xbp-1 genomic DNA in PVD does not (C and D). (E and F) Expressing ER chaperone HSP-4 in PVD rescues the dendritic defect in ire-1 mutants. Anterior, left; dorsal, top. Asterisks, cell bodies. Scale bar, 50 μm. Error bars show mean ± s.e.m., n = 10.
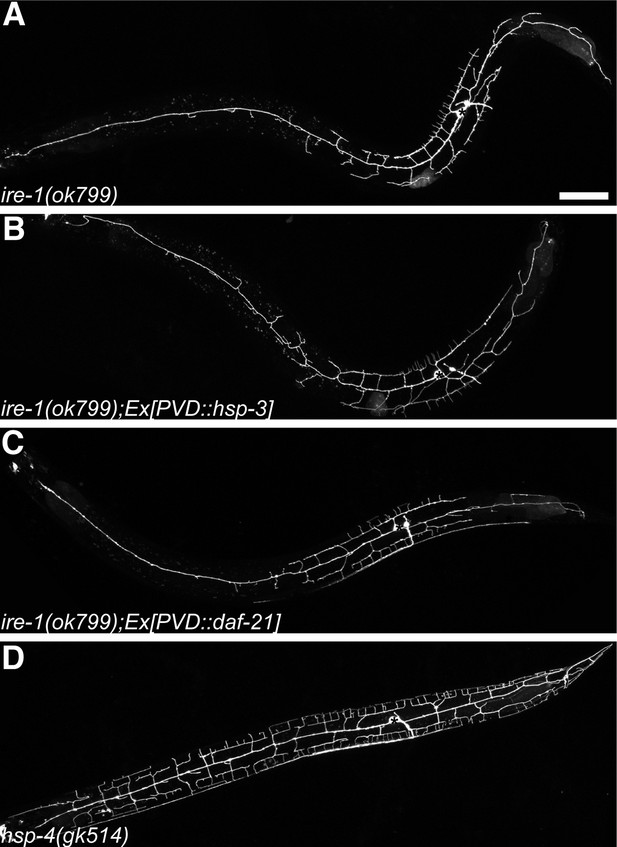
Dendritic morphogenesis defects of PVD is likely due to lack of specific ER chaperones.
Defective dendritic branching in ire-1 mutants (A) cannot be rescued by overexpressing another ER chaperone HSP-3 (B) or a cytoplasmic chaperone DAF-21 in PVD (C). No detectable PVD branching defect in hsp-4(gk514) single mutant (D). Asterisks, cell bodies. Scale bar, 50 μm.

The RIDD pathway in parallel with XBP-1 to regulate PVD dendritic arbor development.
No PVD dendritic arbor defects are shown in xbp-1 null mutant (B) compared to ire-1 mutant (A) while in xbp-1 background, conditional knockout xrn-1 induced by somatic CRISPR phenocopied the subtle ire-1 like phenotype (C). Asterisks, cell bodies. Scale bar, 50 μm.
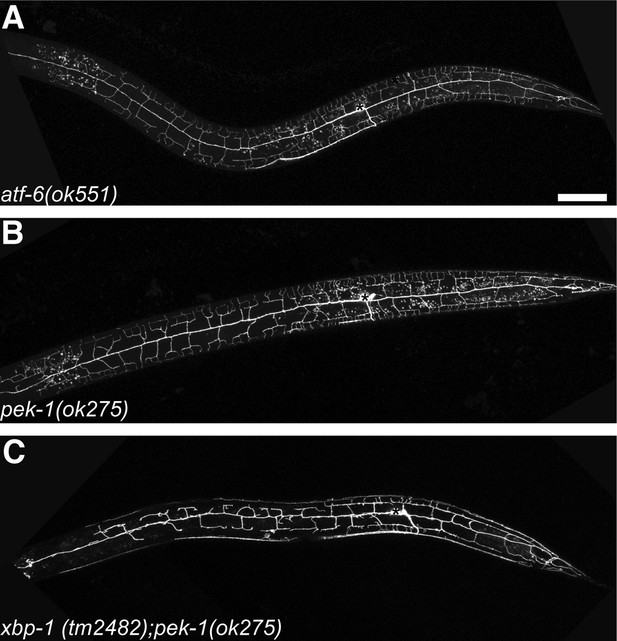
Other UPR arms also contribute to dendrite morphogenesis of PVD.
No PVD dendritic arbor defects was shown in atf-6 (A) or pek-1 (B) null mutants while xbp-1 pek-1 double mutant showed ire-1 like phenotype with low-penetrance (C). Asterisks, cell bodies. Scale bar, 50 μm.
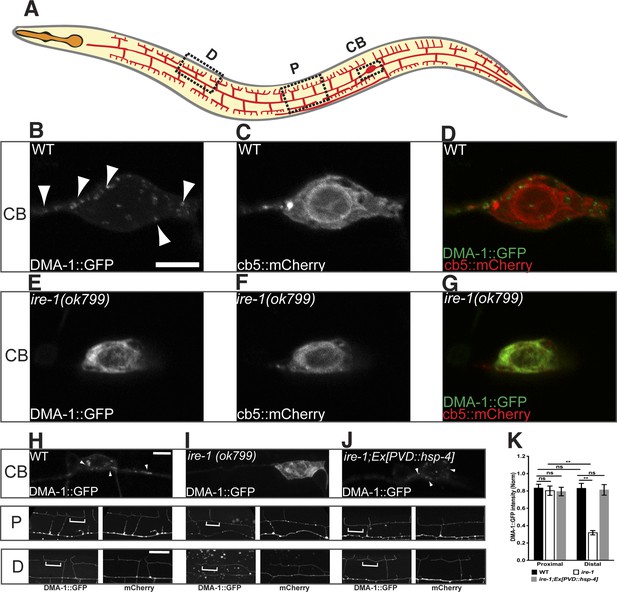
DMA-1 is stuck in the somatic ER in ire-1 mutants.
(A) Diagram of PVD in a young adult animal showing three representative subcellular regions: CB (cell body), P (proximal), and D (distal) dendrites. (B to G) Subcellular localization of DMA-1::GFP and general ER marker cb5::mCherry in PVD cell bodies in wild-type animals (B to D) and ire-1 mutants (E to G). (H to J) DMA-1::GFP subcellular localization in WT (H), ire-1 (I) and ire-1 mutants expressing HSP-4 (J). Top panels: cell body; middle panels: proximal menorahs; bottom panels: distal menorahs. The morphology of the dendritic menorah is shown by cytoplasmic mCherry. Arrowheads, DMA-1::GFP puncta; brackets, tertiary branches (without puncta) used for measuring diffuse DMA-1:GFP. (K) Quantification of diffuse DMA-1:GFP (normalized to cytoplasmic mCherry) on tertiary branches in (H to J). Error bars show mean ± s.e.m., n = 50–60. ns, not significant; **p < 0.01 (two-way ANOVA and post hoc Sidak's multiple comparisons test). Scale bars, 5 μm.
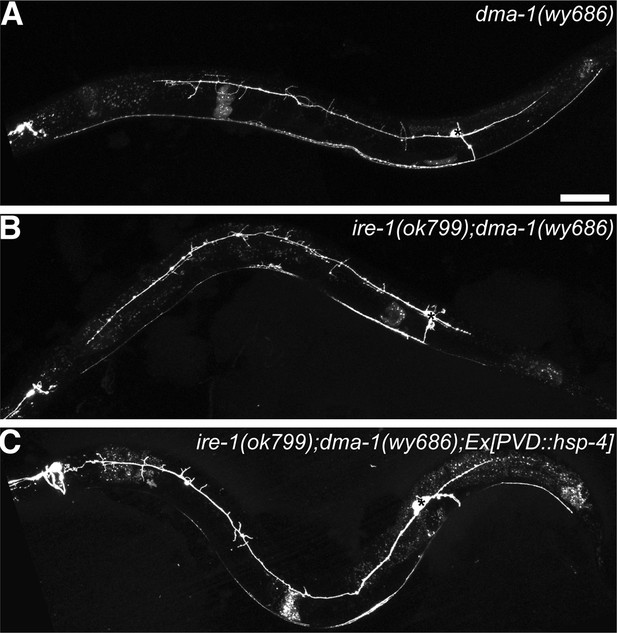
DMA-1 is required for PVD dendrite morphogenesis and may be the downstream of HSP-4.
(A) Severe PVD dendritic branching defect in dma-1 mutants. (B) ire-1;dma-1 double mutants show a similar dendritic phenotype to dma-1 single mutants. (C) Overexpression of hsp-4 in ire-1;dma-1 double mutants cannot rescue the dendritic arbor defect. Asterisks, cell bodies. Scale bar, 50 μm.
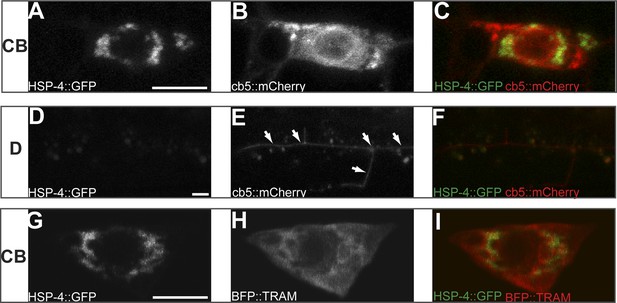
Subcellular localization of HSP-4::GFP, general ER marker cb5::mCherry and rough ER marker BFP::TRAM in PVD cell body (CB) region (A to C and G to I) and in distal (D) dendritic region (D to F) in wild type animals.
Arrows indicating the general ER marker in PVD distal branches. Scale bar, 5 μm.
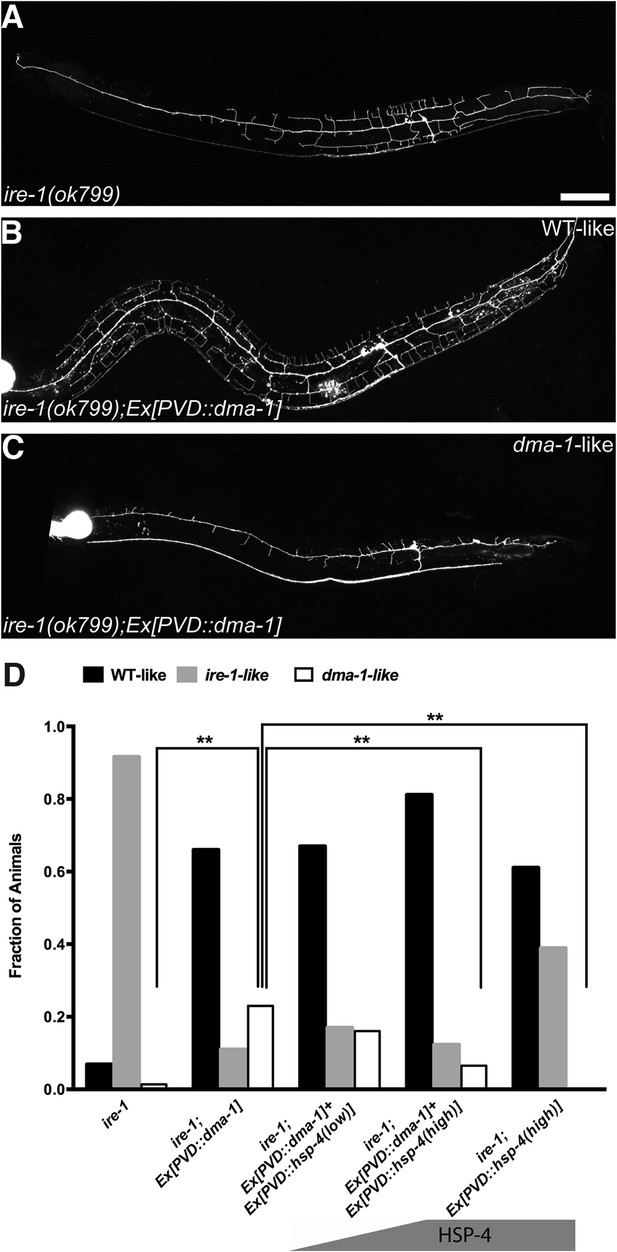
Overexpressing DMA-1 in ire-1 mutants can either rescue dendritic defects or cause more severe dendrite branching defects.
(A) Representative defective PVD dendritic arbor in ire-1 mutants. (B and C) Overexpressing DMA-1 in PVD in ire-1 mutants rescues the dendritic defect (WT-like) (B) or causes a more severe phenotype with fewer branches (dma-1-like) (C). Asterisks, cell bodies; Scale bar, 50 μm. (D) Proportions of different phenotypes in different transgenic rescue strains with overexpression of dma-1 and/or supplementation of different doses of HSP-4 chaperon. n > 120. **p < 0.01, χ2 test with Sidak's multiple comparison correction.
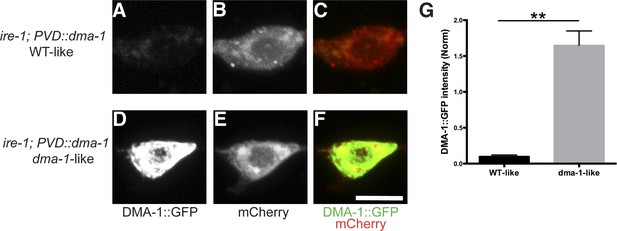
Accumulation or aggregation of DMA-1::GFP in the PVD cell bodies is tightly correlated PVD dendrite morphology.
In the same trangene (wyEx7338) bearing dma-1::GFP expression in ire-1 mutants, there is much less accumulation or aggregation of DMA-1::GFP in the PVD cell bodies with the rescued ‘WT-like’ PVD morphology (A to C) compared with more severely defective ‘dma-1 like’ animals (D to F). (G) Quantification of the DMA-1::GFP intensity in the cell bodies (normalized to cytoplasmic mCherry) in (A to F). Error bars show mean ± s.e.m. n = 20. **p < 0.01, t-test. Scale bar, 5 μm.
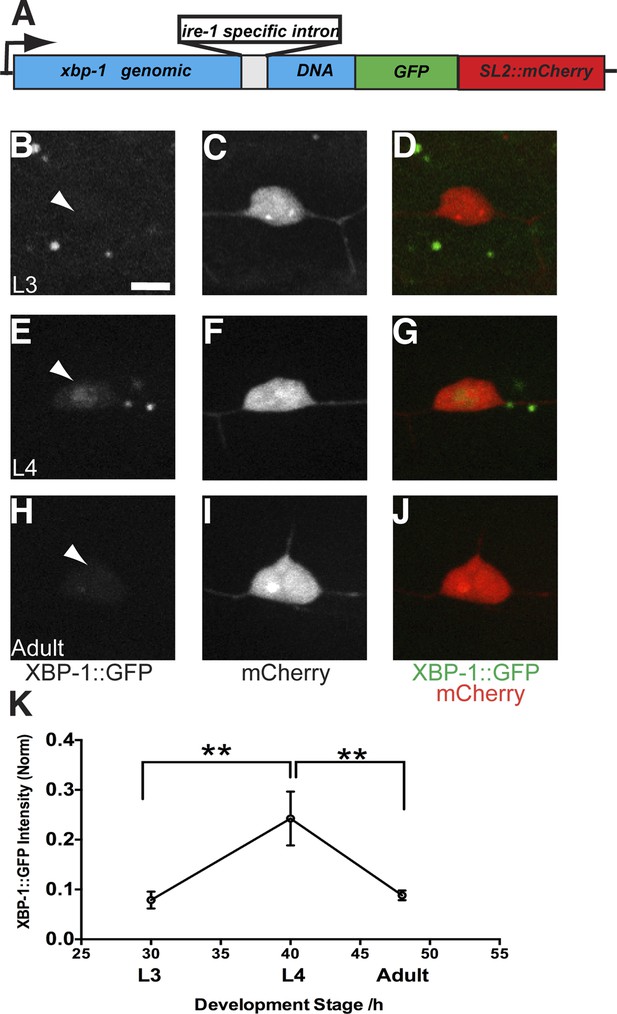
The UPR activity is correlated with dendritic branching during development in the PVD neuron.
(A) Design of the PVD-specific UPR activity reporter. The xbp-1 genomic DNA is fused with GFP followed by an SL2::mCherry cassette, which permits the bicistronic expression of XBP-1::GFP and mCherry. The reporter is driven by a PVD specific promoter. The XBP-1::GFP brightness indicates the UPR activity while the intensity of mCherry is used to normalize to transgene expression levels. (B to J) The PVD UPR activity in L3 stage (B to D), L4 stage (E to G) and adult stage (H to J). Arrowheads, nuclei of PVD. Scale bar, 5 μm. (K) Quantification of the normalized UPR activity in PVD in (B to J). Error bars show mean ± s.e.m. n = 30–50. **p < 0.01, Kruskal–Wallis one-way test and post hoc Dunn's test.

The UPR activity is correlated with dendritic branching during development in the PVD neuron with another UPR reporter.
The UPR activity is indicated by nuclear HIS-24::GFP brightness (driven by the hsp-4 promoter) while the PVD neuron is labeled and identified by cytoplasmic mCherry. The UPR activity of PVD is shown in L3 stage (A to C), L4 stage (D to F) and adult stage (G to I). Arrowheads, nuclei of PVD. Scale bar, 5 μm. (J) Quantification of the UPR activity in PVD in (A to I). a.u., arbitrary unit. Error bars show mean ± s.e.m. **p < 0.01, Kruskal–Wallis one-way test and post hoc Dunn's test.
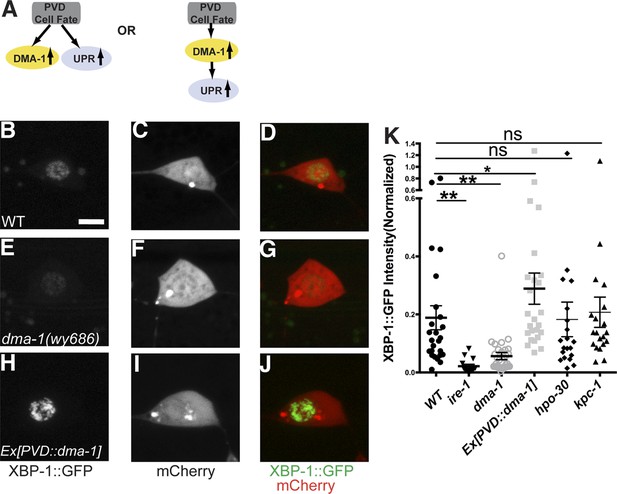
The induction of the UPR in PVD depends on the DMA-1.
(A) Diagrams showing two possible models for the activation of the UPR in PVD. (B to J) The PVD UPR activity in WT (B to D), dma-1 mutants (E to G) and WT with overexpession of dma-1 (H to J). Scale bar, 5 μm. (K) Quantification of the normalized UPR activity in PVD in (B to J) and other mutants in (Figure 6—figure supplement 1D–L). Error bars show mean ± s.e.m. ns, not significant, **p < 0.01, *p < 0.05, Kruskal–Wallis one-way test and post hoc Dunn's test.
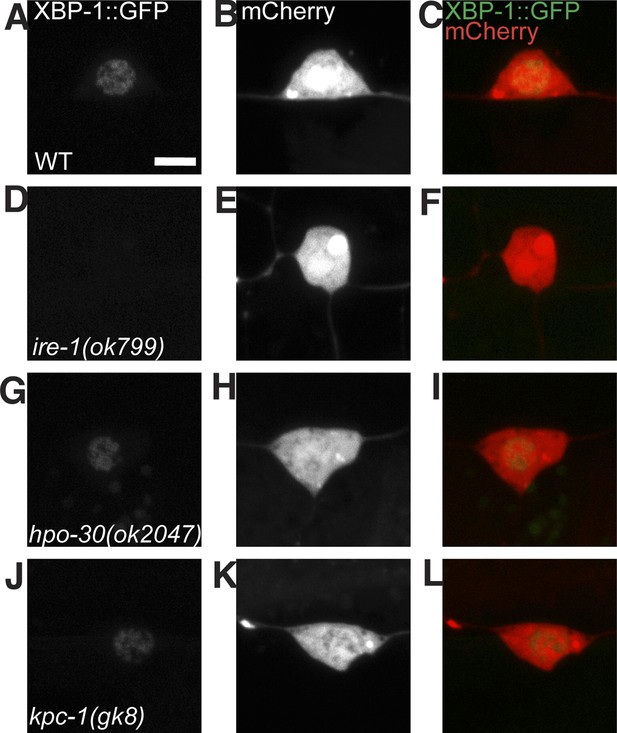
The UPR activity in PVD does not depend on other known proteins that are processed in the ER and are required for PVD dendrite morphogenesis.
The PVD UPR activity in WT (A to C), ire-1 (D to F), hpo-30 (G to I) and kpc-1 mutants (J to L). UPR activity is indicated by XBP-1::GFP brightness while the intensity of mCherry is used to normalize for transgene expression level. Scale bar, 5 μm.

Another UPR reporter in PVD also showed dramatic decrease in dma-1 mutants.
The UPR activity is indicated by nuclear HIS-24::GFP brightness (regulated by hsp-4 promoter) while the intensity of cytoplasmic mCherry is used to normalize for transgene expression level. The UPR activity of PVD is shown in WT (A to C), ire-1 (D to F) and dma-1 (G to I) animals. Arrowheads, nuclei of PVD. Arrow, PDE cell body occasionally labeled in this strain. Scale bar, 5 μm. (J) Quantification of the normalized UPR activity in PVD in (A to I). Error bars show mean ± s.e.m. **p < 0.01, Kruskal–Wallis one-way test and post hoc Dunn's test.
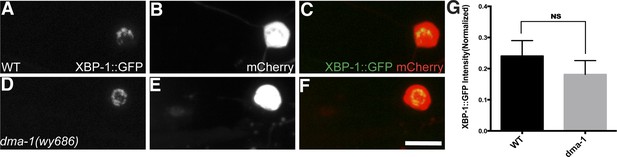
The UPR in unbranched PVC neurons does not depend on DMA-1.
The UPR activity in WT (A to C) and dma-1 mutants (D to F). UPR activity is measured by XBP-1::GFP brightness while the intensity of mCherry is used to normalize for transgene expression level. Scale bar, 5 μm. (G) Quantification of the normalized UPR activity in PVC in (A to F). Error bars show mean ± s.e.m. n = 20. NS, not significant, t-test.
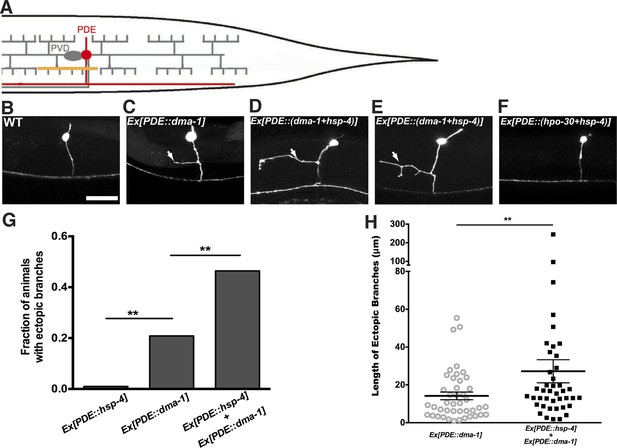
Expression of dma-1 and hsp-4 together in the morphologically simple PDE neurons can induce ectopic branching more dramatically.
(A) Diagram showing the PDE neuron (in red) which is located close to the PVD cell body and has a simple processes running adjacent to the PVD dendrites. Orthogonal ectopic branching (in orange) at a stereotyped position in the PDE commissure, reminiscent in location and direction to PVD tertiary branching. (B to D) Representative dendritic morphology of PDE neurons expressing cytoplasmic GFP in wild-type (B), a strain with expression of dma-1 (C) and a strain with co-expression of dma-1 and hsp-4 in PDE (D). Scale bar, 20 μm. Arrows, orthogonal ectopic branches. (E) More dramatic ectopic branching in the same strain co-expressing dma-1 and hsp-4 in PDE. (F) No ectopic branching in the same strain co-expressing hpo-30 and hsp-4 in PDE. (G) Percentages of PDE ectopic branching in strains expressing hsp-4 only, dma-1 only, and dma-1 with hsp-4. n > 100. **p < 0.01, χ2 test with Sidak correction. (H) Length of PDE ectopic branches in the strains expressing dma-1 only, and dma-1 with hsp-4. **p < 0.01, Mann Whitney U-test.

The defective PVD phenotype of ire-1 is suppressed by reduced size of PVD dendritic arbor.
(A). Defective dendritic branch in ire-1 mutants (B). Reduced body length and smaller PVD size in dpy-5 mutants. (C). dpy-5 ire-1 double mutants showing partially rescued PVD morphology. (D). Some dpy-5 ire-1 double mutants showing wild-type PVD morphology. (E). Quantification of numbers of secondary branches per 100 μm in (A–D). Asterisks, cell bodies. Scale bar, 50 μm. **p < 0.01, one-way ANOVA and post hoc Sidak's multiple comparisons test.

Overexpression of dma-1 in PDE induces ectopic branches and increases UPR activity.
Representative morphology of PDE neurons expressing cytoplasmic mCherry in wild-type (A) and a strain with ectopic expression of dma-1 in PDE (D). In these animals, the UPR activity is indicated by Phsp-4::HIS-24::GFP reporter in (B) and (E). Asterisks, PDE nuclei. Arrow, orthogonal ectopic branch of PDE. Scale bar, 5 μm. (G) Quantification of the UPR activity in PDE in (A to F). a.u., arbitrary unit. Error bars show mean ± s.e.m. **p < 0.01, Mann Whitney U-test.
Additional files
-
Source code 1
Custom built software in Matlab and Python codes. Zip file contains: findPrimary.m to find the PVD primary dendrite, and it calls BDB function (Peng, H, Long, F, Liu, X, Kim, SK, and Myers, EW (2008). Straightening Caenorhabditis elegans images. Bioinformatics 24, 234–242. Cited paper in the manuscript), which is available from http://penglab.janelia.org/proj/wormatlas/bdb_minus_demo_download.html. SplitWorm.m to split the primary dendrite into equal length fragments. PrintSplits.m to draw lines to indicate the segmentation of the primary dendrite. ImageProcessing.py: the python code for image processing and data analysis.
- https://doi.org/10.7554/eLife.06963.025





