miR-965 controls cell proliferation and migration during tissue morphogenesis in the Drosophila abdomen
Figures
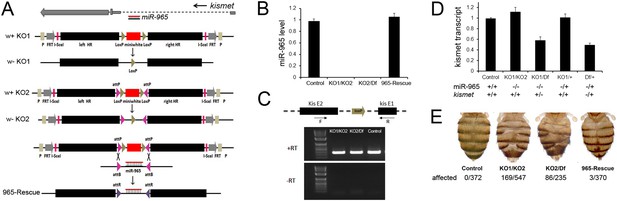
The miR-965 mutant.
(A) miR-965 is located within the first intron of the kismet gene. The targeting strategies used to produce two independent miR-965 deletion mutants by ends-out homologous recombination are shown below. Left and Right homology arms cloned into in the targeting vector are shown in black. The w+ KO1 mutant was made by replacing miR-965 with a mini-white reporter (red) flanked by LoxP sites (grey). Sections are not represented to scale. w-KO1 indicates the targeted allele after Cre-mediated excision of the mini-white cassette. The w+ KO2 mutant was made by replacing miR-965 with mini-white flanked by LoxP sites and inverted attP sites (pink). w-KO2 indicates the allele after Cre-mediated excision of mini-white. Use of RMCE to replace the mini-white cassette with the miRNA to produce the 965-Rescue allele is shown at bottom. (B) miR-965 RNA level measured by quantitative miRNA PCR. RNA was isolated from adult flies of the indicated genotypes. Control was w1118. Df indicates Df(2L)ED19. Data represent the average of 3 independent experiments ± standard deviation (SD). (C) RT-PCR using primers flanking the first intron of kismet. A PCR product of normal size was produced using RNA from flies of each of the indicated genotypes. No product was produced in the absence of reverse transcriptase. (D) Quantitative real-time RT-PCR showing kismet transcript levels in miR-965 mutants (KO1/KO2 and KO1/Df) and the heterozygous KO1/+ and Df/+ controls. Data represent the average of 3 independent experiments ± SD. (E) Dorsal aspect of the abdomen from females with the indicated mutant combinations. Control was w1118. The number of affected individuals is shown below. ANOVA: p < 0.0001 for each mutant genotype compared to the w1118 control or to the rescued mutant.
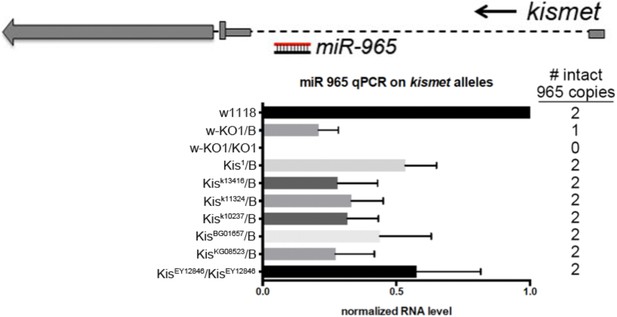
Evidence that kismet and miR-965 arise from a common transcription unit.
Above: diagram of the kismet locus showing miR-965 in the first intron. kismet and miR-965 are transcribed in the same direction. Below: quantitative miRNA PCR showing the level of miR-965 miRNA. miR-965 levels were reduced in flies carrying several kismet alleles associated with different transgene insertions near the first exon, in which both copies of the miRNA gene should be intact. Genotypes are shown at left and the expected number of copies of miR-965 DNA is shown at right. miR-965 levels were somewhat lower than the expected 50% in the KO/+ heterozygote. ‘B’ indicates balancer chromosome. Refers to Figure 1A–D.

Phenotype classification.
Images showing the three classes of defect: gaps, or lack of tissue; fusion, and polarity reversal. Right panels show higher magnification views of the bristle pattern to illustrate the polarity reversal phenotype. Refers to Figure 1E.
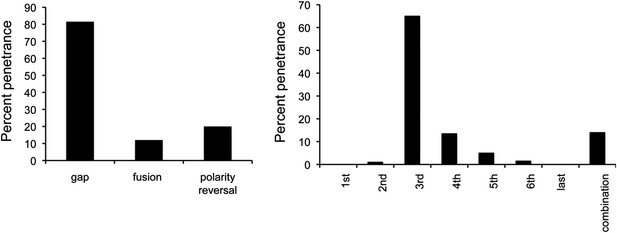
Penetrance of defects in miR-965 mutants, shown as % of affected individuals.
Large gaps associated with fusion of adjacent segments occurred in ∼10% of cases. Gap penetrance per segment is shown at right. n = 200 flies. Refers to Figure 1E.
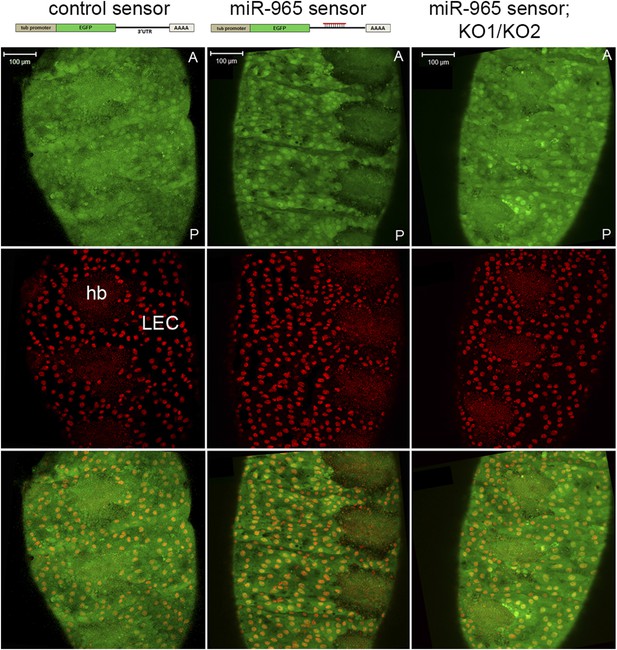
miR-965 expression in histoblasts.
Top: design of the control and miR-965 sensor transgenes. EGFP was under control of the tubulin promoter. For the miR-965 sensor, 1 copy of a perfect miR-965 target sequence was placed into the SV40 UTR. Images showing GFP expression from the control sensor (left) and miR-965 sensor (middle) transgenes at 21 hr APF. Histoblast nests consist of small diploid histoblast cells (hb) surrounded by large polyploid larval epidermal cells (LEC). Nuclei were labeled with histone-RFP (red). Downregulation of GFP was lost when the transgene was placed in the KO1/KO2 miR-965 mutant background (right). Anterior (A), posterior (P). Scale bar: 100 µm.
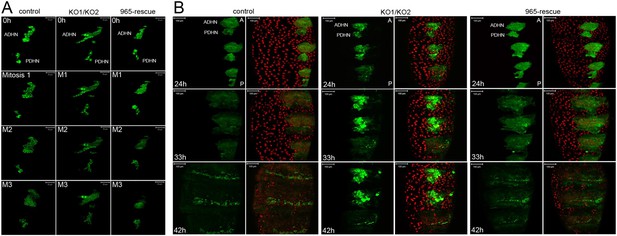
Abnormal histoblast proliferation and migration in the miR-965 mutant.
(A) Still images taken from time-lapse videos of control, miR-965 mutant (KO1/KO2) and rescued mutant showing the reduction divisions of the early histoblast proliferation phase. M1, M2 and M3 indicate images taken after mitosis 1, 2 or 3. Imaging was started 0–1 hr APF. Histoblasts were labeled by esg-Gal4 directed expression of UAS-nuclear GFP. ADHN and PDHN represent anterior dorsal histoblast nests and posterior dorsal histoblast nests. Scale bars: 50 µm. Note the different cell sizes in the miR-965 mutant histoblast nests. (B) Still images taken from time-lapse videos at 24, 33 and 42 hr APF from control, miR-965 mutant and rescued mutant to illustrate expansion of the histoblast nests to replace LECs. Histoblasts were labeled by esg-Gal4 directed expression of cytoplasmic GFP. esg-GAL4 and UAS-GFP were recombined onto the miR-965 mutant and onto the miR-965 Rescue chromosome. Nuclei were labeled red with H2-RFP A and P indicate anterior and posterior orientation. Scale bars: 100 µm.
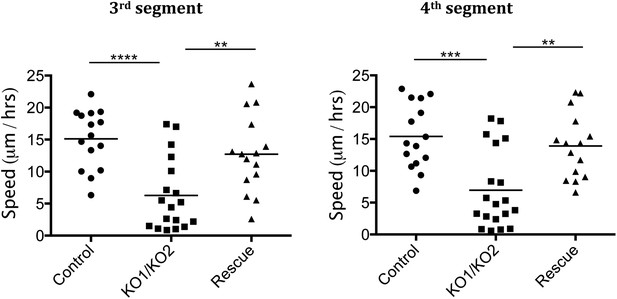
Rate of histoblast nest expansion measured from time-lapse videos.
Each data point corresponds to one histoblast nest. The leading edge of each histoblast nest was tracked using imageJ. Speed was calculated measuring total distance covered (micrometer/hour). Genotypes: Control was esg-GAL4, UAS-GFP. esg-GAL4 and UAS-GFP were recombined onto the KO1 and KO2 chromosomes and onto the miR-965-Rescue chromosome. Data include examples with both recombinant mutant chromosomes. No difference between these two recombinants was apparent. n = 18 for the miR-965 (KO1/KO2) mutant combination. n = 15 for control and rescue. Left panel: p < 0.0001 comparing KO1/KO2 with control, p < 0.01 comparing KO1/KO2 with rescue using one-way ANOVA. Right panel: p < 0.001 comparing KO1/KO2 with control, p < 0.01 comparing KO1/KO2 with rescue using one-way ANOVA. Refers to Figure 3B and Videos 5–7.
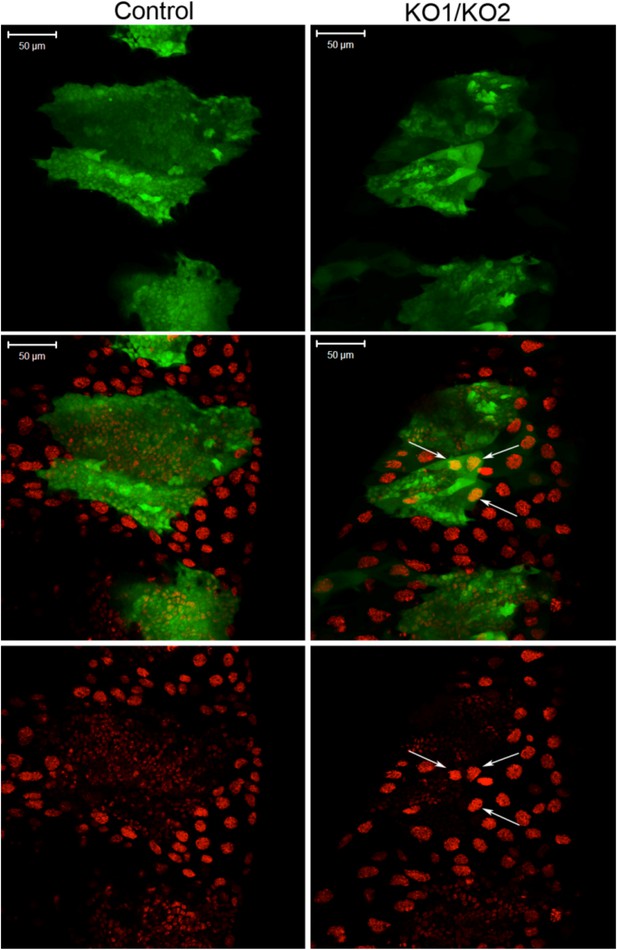
Large polyploid cells in miR-965 mutant histoblast nests.
Histoblast nests were labeled with esg-Gal4-directed expression of UAS-GFP at 24 hr APF. Note the presence of large polyploid cells in the histoblast nest in the miR-965 mutant (arrows). At the start of the imaging period, the large polyploid cells marked by the arrows did not express GFP, but began to express GFP after making contact with the expanding histoblast nests. Possible explanations for the appearance of GFP in large polyploidy cells include (1) induction of esg-Gal4 activity in the larval cells that cannot be eliminated by the expanding histoblast nests, perhaps by signals from the histoblasts; (2) fusion of polyploidy LEC with esg-Gal4-expressing histoblasts. Scale bar: 50 µm. Refers to Figure 3B.
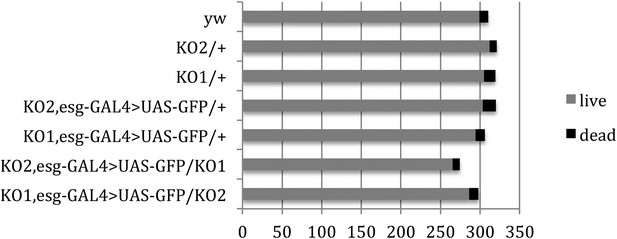
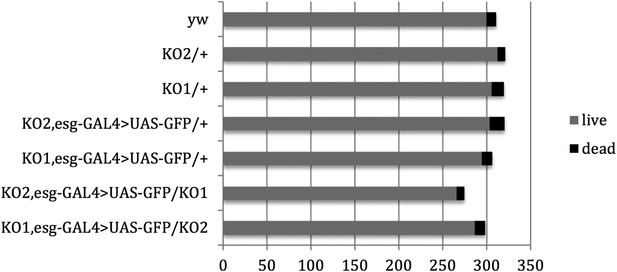
Pupal survival assays.
Pupal survival was assayed for flies of the indicated genotypes. 6 batches of pupae were sampled/genotype. The data present the total number of surviving adults (live) and the total number of dead pupae (dead). There was no significant difference between the mutant and control genotypes used to make the videos: p = 0.67 comparing KO2 esgG4>GFP/+ vs KO2 esgG4>GFP/KO1 (Mann–Whitney test). p = 1 comparing KO1 esgG4>GFP/+ vs KO1 esgG4>GFP/KO2 (Mann–Whitney test). Refers to Figure 3B.
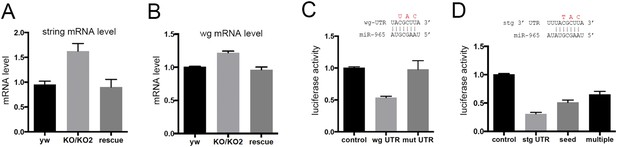
miR-965 regulates string and wingless.
(A, B) string (stg) and wingless (wg) transcript levels measured by quantitative real time RT-PCR in RNA isolated from w1118 control, KO1/KO2 and 965-rescue pupae at 21 hr APF. Data represent the average of three independent RNA collections ± SD. ANOVA: p < 0.01 comparing KO1/KO2 with control or with rescue for stg and wg. (C) Top: diagram of the predicted miR-965 target site in the wg 3′ UTR, showing pairing to the miRNA seed sequence. Residues shown in red were mutated in the mutant version of the wg 3′ UTR luciferase reporter. Below: luciferase activity in S2 cells transfected to express a tubulin-promoter miR-965 transgene, Renilla luciferase and the indicated firefly luciferase reporters. Control indicates the luciferase reporter with the SV40 3′ UTR, which lacks miRNA binding sites. wg UTR indicates the intact full-length wg 3′ UTR. Mut indicates the wg 3′ UTR with the miRNA seed site mutated as indicated in red. Data represent the average of 3 independent experiments ± SD. ANOVA: p < 0.001 comparing control to the intact 3′ UTR. p = 0.001 comparing the intact and site mutant versions of the 3′ UTR. (D) Top: diagram of the predicted miR-965 target site in the stg 3′ UTR, showing pairing to the miRNA seed. Residues shown in red were mutated in the seed mutant version of the reporter. The changes made in the extended target site mutant reporter are shown in Figure 3. Below: luciferase activity as in panel C. Data represent the average of 3 independent experiments ± SD. ANOVA: p < 0.0001 comparing control to the intact 3′ UTR and comparing intact to seed mutant and multiple mutant UTR reporters.
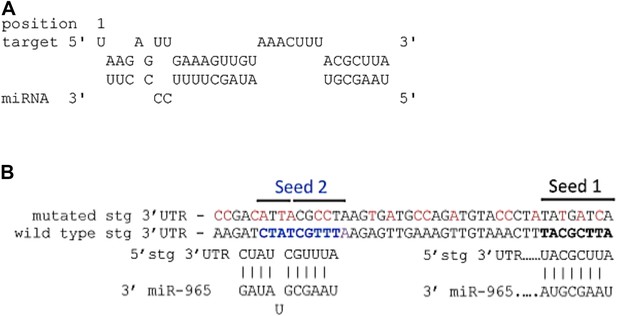
(A) Predicted miR-965 sites in the string 3′UTR.
Based on the potential for strong 3′ pairing in the Seed 1 mutant (shown in Figure 4D), as well as the presence of a second nearby non-canonical seed match (seed 2), a more extensively mutated UTR was made to eliminate pairing to both potential sites. Nucleotides mutated are shown in red. Refers to Figure 4D. (B) Structure of the miR-965 site in the string 3′ UTR, as predicted by RNAHybrid (http://bibiserv.techfak.uni---bielefeld.de/).
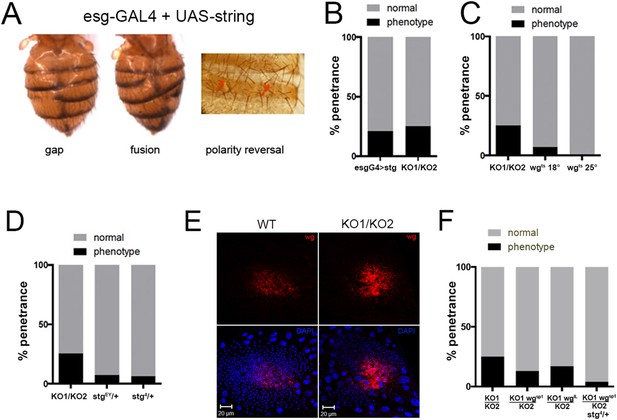
Overexpression of string and wg contributes to the miR-965 mutant phenotype.
(A) Dorsal views of abdomens from adult female esg-Gal4 UAS-string flies illustrating the segment gap, segment fusion and polarity reversal phenotypes. (B) Penetrance of abdominal defects of all classes in esg-Gal4 UAS-string vs mutant. esg-Gal4 UAS-string: n = 97/469; KO1/KO2 n = 110/446. p = 0.16 Fishers exact test. (C) Penetrance of abdominal defects in esg-Gal4 UAS-wgts flies reared at 18° and 25°C vs KO1/KO2. esg-Gal4 UAS-wgts reared at 18°C: n = 9/129; esg-Gal4 UAS-wgts at 25°C n = 1/254; KO1/KO2 n = 110/446. p = 0.014 comparing wgts at 18 vs 25°C, Fishers exact test. (D) Penetrance of abdominal defects comparing KO1/KO2 mutants with KO1/KO2 mutants carrying one copy of stringEY12388 or string4 alleles. p < 0.001 comparing KO1/KO2 to KO1/KO2; stgEY/+ or stg4/+ using Fisher's exact test. (E) Confocal micrographs showing dorsal histoblast nests of wild-type (WT) and miR-965 mutant (KO) at ∼24 hr APF labeled with anti-Wg (red). Nuclei were labeled with DAPI (blue). Scale bar: 20 µm. Anterior and dorsal histoblast nests in the miR-965 mutants were not yet fused at 24 hr APF, due to delayed migration. Images were captured using identical microscope settings. (F) Penetrance of abdominal segmentation defects comparing KO1/KO2 mutants with KO1/KO2 mutants carrying one copy of wgSP-1 or wgl-12 temperature sensitive alleles or carrying one copy of wgSP-1 and stg4 together. p < 0.05 comparing KO1/KO2 to KO1, wgSP-1/KO2 using Fisher's exact test. KO1/KO2 was not significantly different from KO1, wgI-12/KO2, perhaps because wgI-12 is a weaker, temperature sensitive allele. p < 0.001 comparing KO1/KO2 with KO1, wgSP-1/KO2; stg4/+ using Fisher's exact test.
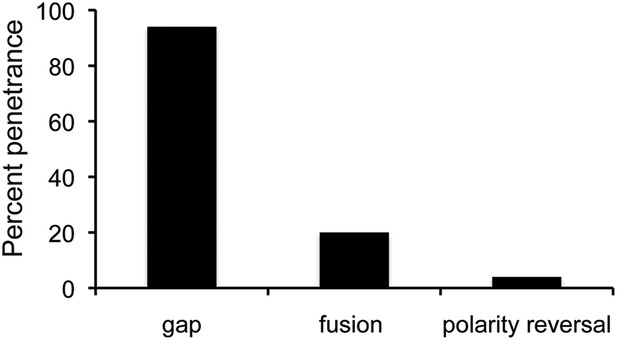
The proportion of flies with defects caused by string overexpression.
Penetrance of the three types of abdominal defect in flies overexpressing UAS-String under esg-Gal4 control. n = 102 flies. Refers to Figure 5A.
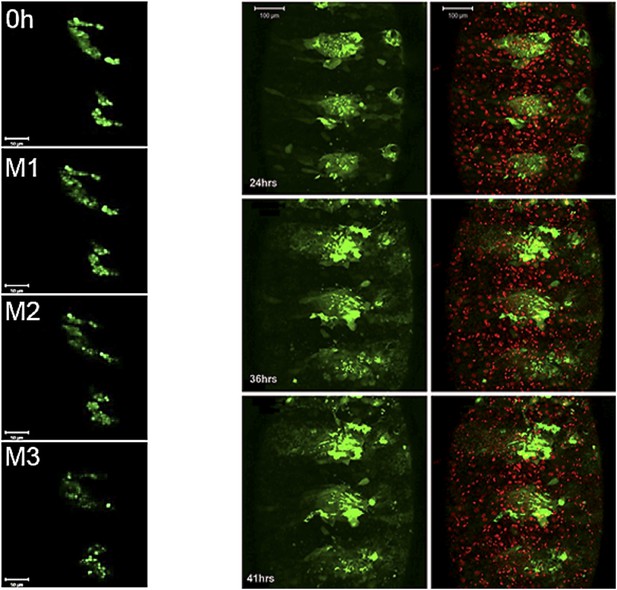
Still images from a time-lapse video of esg-Gal4>UAS-string histoblasts.
Left: rapid proliferation phase. Note the presence of cells of different sizes, indicative of asynchronous division. Images represent 0 hr and mitosis M1, M2 and M3. Genotype: esg-Gal4, UAS-string, UAS-GFP. Right: growth and migration phase. Note the delayed spreading and incomplete replacement of the LEC, compared to controls at the equivalent time points (Figure 3C). Genotype: esg-Gal4, UAS-string, UAS-GFP. Refers to Figure 5B and Videos 8, 9.
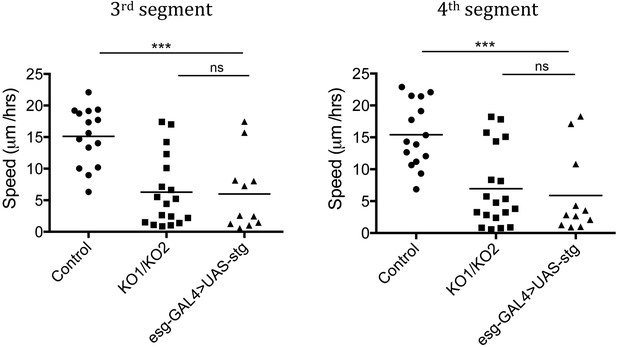
Speed of histoblast nest migration.
n = 15 for control, n = 18 for KO1/KO2 and n = 11 for esg-GAL4>UAS-stg. p < 0.001 comparing control and esg-GAL4>UAS-stg using one-way ANOVA. esg-GAL4>UAS-stg is not significantly different from the miR-965 mutant (KO1/KO2). Control and miR-965 mutant samples are same as in Figure 3—figure supplement 1. Refers to Figure 5B and Video 9.
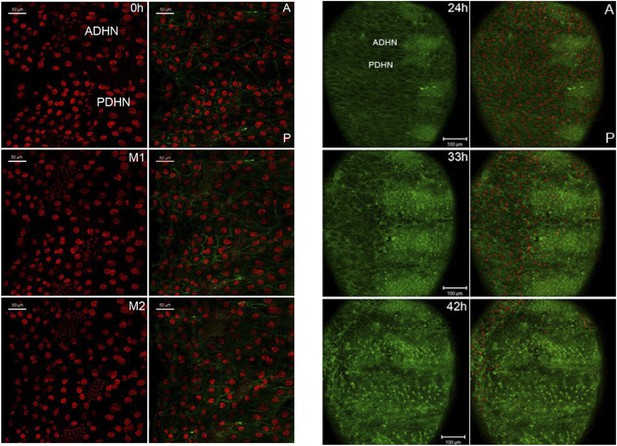
Rescue of the migration defect of miR-965 mutants with reduced levels of string.
Left: still images taken from a time-lapse video showing histoblast divisions in the miR-965 mutant (KO1/KO2) with reduced levels of string transcript using the stgEY12388 allele. M1 and M2 indicate mitosis 1 and 2. Cell membranes were labeled using Atpα-GFP (green). Nuclei were labeled using Histone2-RFP (red). ADHN: anterior dorsal histoblast nest. PDHN: posterior dorsal histoblast nest. Scale bars: 50 µM. Right: still images taken from a time-lapse video showing histoblast nest migration in animals of the same genotypes. Scale bars: 100 µM. Refers to Figure 5D and Video 11.
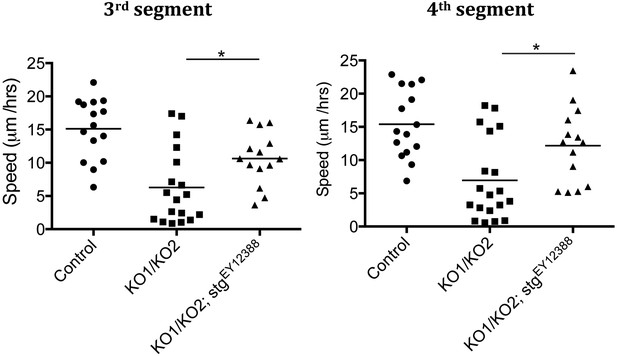
Speed of histoblast migration restored by reduced string activity.
Left: speed of histoblast nest migration in the third abdominal segment. p < 0.05 comparing KO1/KO2 with KO1/KO2; stgEY12388. Control (n = 15), KO1/KO2 (n = 18) and KO1/KO2; stgEY12388 (n = 14). The Control and KO1/KO2 samples are the same as those in Figure 3—figure supplement 1. The two experiments were done together. Right: speed of histoblast migration in the fourth abdominal segment. p < 0.05 comparing KO1/KO2 with KO1/KO2; stgEY12388. Refers to Figure 5D and Video 11.
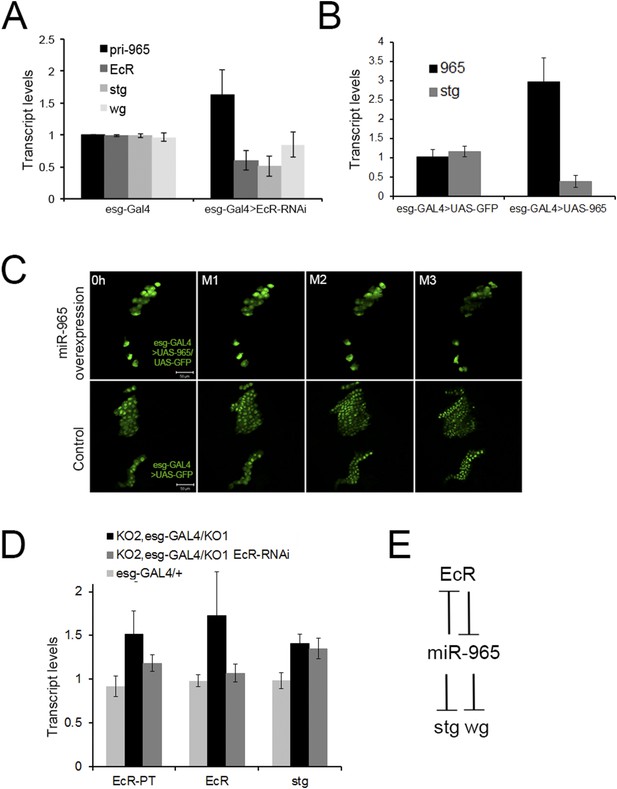
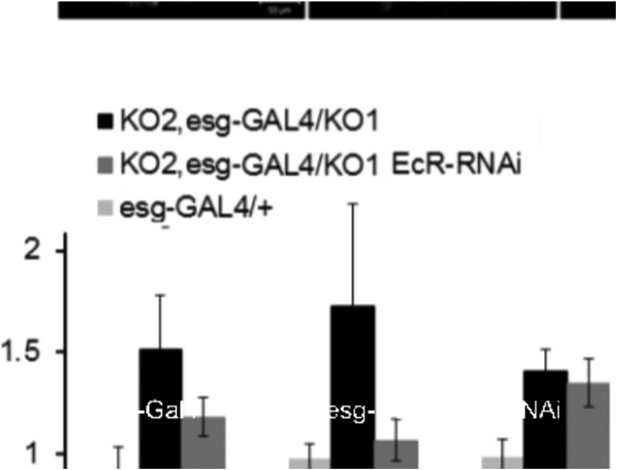
Regulation of miR-965 by ecdysone at the beginning of pupariation.
(A) Quantitative RT-PCR showing levels of miR-965 primary transcript, EcR, and string mRNAs in RNA isolated from pupae expressing esg-GAL4 (control) and esg-GAL4 driving UAS-EcR-RNAi to deplete EcR mRNA. Samples were collected at 0 hr APF. Data were normalized to rp49 and to the esg-GAL4 control. Data represents average of three independent samples ± SD. (B) Quantitative RT-PCR showing string mRNA in 0 hr pupae overexpressing miR-965 in histoblast cells. For quantitative microRNA PCR, data were normalized to U14, U27, SnoR422. Data were normalized to rp49 for string mRNA qPCR. Data represent the average of four independent samples ± SD. (C) Images from time-lapse videos showing the effects of miR-965 overexpression in histoblast cells during the synchronous division phase. M1, M2 and M3 indicate three consecutive mitotic divisions in dorsal histoblast nests. Scale bar: 50 µm. (D) Quantitative RT-PCR showing levels of string, EcR primary transcript (EcR-PT) and mature mRNA in RNA isolated from pupae expressing esg-GAL4 (control), esg-GAL4 in the miR-965 mutant with and without UAS-EcR-RNAi to deplete EcR mRNA. esg-GAL4 was recombined onto the KO2 mutant chromosome. Samples were collected at 0 hr APF. Data were normalized to rp49 and to the esg-GAL4 control. Data represents average of six independent samples ± SD. p = 0.37 for stg levels between KO2, esg-GAL4/KO1 and KO2, esg-GAL4/KO1>EcR-RNAi. p ≤ 0.01 comparing primary and mature EcR transcripts between esg-GAL4 control and KO2, esg-GAL4/KO1 mutant samples. (E) Diagram of the regulatory relationships between EcR, miR-965 and the miR-965 targets string and wg. The symbols represent repression of gene expression. miR-965 and EcR repress each other at the primary transcript level. The effect of miR-965 on EcR primary transcript is most likely indirect.
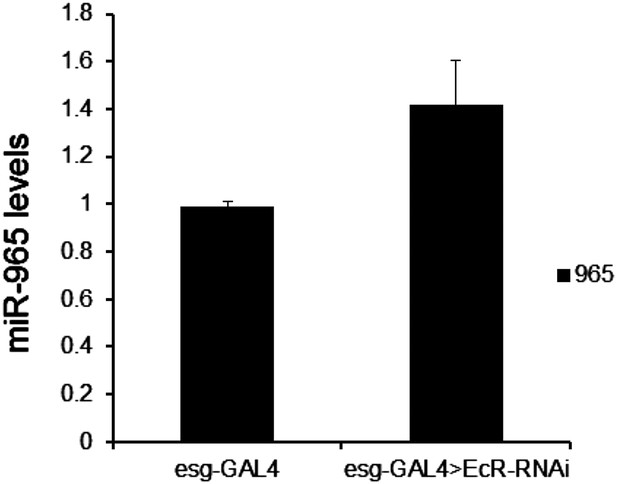
Mature miR-965 miRNA regulation by EcR.
miRNA quantitative RT-PCR showing the levels of mature miR-965 in RNA extracted from 0 hr pupae. esg-GAL4 was used to direct UAS-EcRRNAi expression in histoblasts. Data were normalized to U27 and snoR422 and to the esg-GAL4/+ control sample. Data represent the average of three independent biological samples ± SD. Refers to Figure 6A.
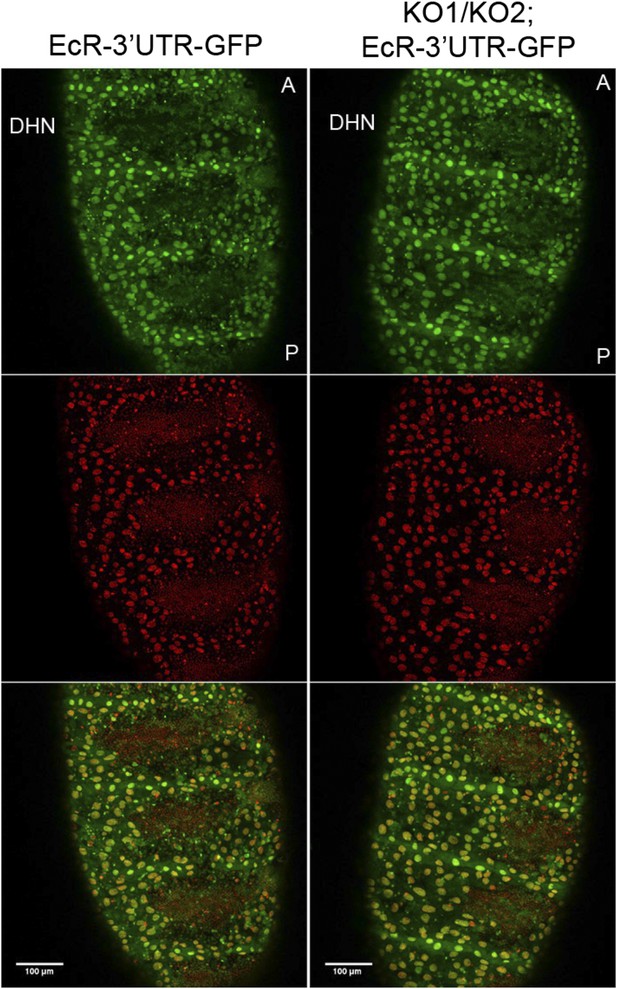
EcR 3′ UTR reporter expression in the miR-965 mutant.
A reporter transgene containing the n EcR 3′ UTR linked to GFP was introduced into the miR-965 KO1/KO2 mutant background. GFP expression (green) did not increase in the histoblast nests in the miRNA mutant compared to the control. Thus, there was no indication that the miRNA acts directly on EcR transcript. There were no good quality miR-965 sites predicted in the various EcR 3′ UTR isoforms. Nuclei were labeled with H2-RFP. Scale bar—100 µM. Refers to Figure 6D.
Videos
Control, division phase.
Early synchronous divisions of histoblast nests in Esg-GAL4>UAS-nuclear GFP controls. GFP was used to track anterior and posterior dorsal histoblast nests during the early division phase. Animals were collected for imaging at 0 hr APF, the white pre-pupal stage. ADHN: Anterior dorsal histoblast nest. PDHN: posterior dorsal histoblast nest. A and P indicate anterior and posterior orientation. Scale bar: 50 µM. Refers to Figure 3A.
miR-965 mutant division phase.
Early asynchronous divisions in the miR-965 mutant (KO2, esg-GAL4>UAS-nuclear GFP/KO1). Esg-GAL4>UAS-nuclear GFP was recombined onto miR-965 RMCE mutant (KO2) chromosome. ADHN and PDHN indicate anterior and posterior dorsal histoblast nests. Animals were collected for imaging at 0 hr APF. Scale bar: 50 µM. Refers to Figure 3A.
miR-965 mutant apoptosis.
Apoptotic cells are seen during the early histoblast division phase in the miR-965 mutant. Esg-GAL4>UAS-nuclear GFP was recombined onto miR-965 RMCE mutant (KO2) chromosome. ADHN and PDHN indicate anterior and posterior dorsal histoblast nests. Animals were collected for imaging at 0 hr APF. Scale bar: 50 µM. Refers to Figure 3A.
miR-965-Rescue division phase.
Early synchronous divisions of histoblasts in miR-965 RMCE rescue. Esg-GAL4>UAS-nuclear GFP was recombined onto miR-965 RMCE rescue chromosome. ADHN and PDHN indicate anterior and posterior dorsal histoblast nests. Animals were collected for imaging at 0 hr APF. Scale bar: 50 µM. Refers to Figure 3A.
Control, growth phase.
Migration of histoblast nests during the growth phase in an Esg-GAL4>UAS-cytoplasmic GFP control pupa. GFP (green) is used to monitor growth and migration of histoblast nests. H2-RFP (red) marks the nuclei. Big nuclei are LECs and small nuclei are histoblast cells. ADHN and PDHN indicate anterior and posterior dorsal histoblast nests. Scale bar: 100 µM. Refers to Figure 3—figure supplement 1.
miR-965 mutant growth phase.
Delayed migration of histoblast nests during the growth phase in the miR-965 mutant. Esg-GAL4>UAS-cytoplasmic GFP was recombined onto both KO1 and KO2 mutant chromosomes. This video shows a KO2, esg-GAL4>UAS-cytoplasmic GFP/KO1 pupa. H2-RFP (red) marks the nuclei. ADHN and PDHN indicate anterior and posterior dorsal histoblast nests. Scale bar: 100 µM. Refers to Figure 3—figure supplement 1.
miR-965 Rescue growth phase.
Migration of histoblast nests during the growth phase in the miR-965 RMCE rescue genotype. Esg-GAL4>UAS-cytoplasmic GFP was recombined onto the miR-965 RMCE rescue chromosome. H2-RFP (red) marks the nuclei. ADHN and PDHN indicate anterior and posterior dorsal histoblast nests. Scale bar: 100 µM. Refers to Figure 3—figure supplement 1.
Esg-Gal4 UAS-Stg division phase.
Early divisions in a pupa overexpressing String under esg-GAL4 control (genotype: esg-GAL4, UAS-nuclear GFP/UAS-stg). ADHN and PDHN indicate anterior and posterior dorsal histoblast nests. Animals were collected for imaging at 0 hr APF. Scale bar: 50 µM. Refers to Figure 5—figure supplement 2.
Esg-Gal4 UAS-Stg growth phase.
Growth and migration phase in a pupa overexpressing String under esg-GAL4 control, as in Video 8. ADHN and PDHN indicate anterior and posterior dorsal histoblast nests. Scale bar: 100 µM. Refers to Figure 5—figure supplements 2, 3.
miR-965 mutant with reduced string levels division phase.
The string mutant allele, stgEY12388 was used to reduce string levels in the miR-965 mutant (KO1/KO2) background. Atpα-GFP (green) was used to mark cell membranes. H2-RFP (red) marks the nuclei. ADHN and PDHN indicate anterior and posterior dorsal histoblast nests. Scale bar: 50 µM. Refers to Figure 5D.
miR-965 mutant with reduced string levels with reduced string levels growth phase.
Normal growth and migration of histoblast nests in the miR-965 mutant with reduced string levels. The string mutant allele, stgEY12388 was used to reduce string levels in the miR-965 mutant (KO1/KO2) background. Atpα-GFP (green) was used to mark cell membranes. H2-RFP (red) marks the nuclei. ADHN and PDHN indicate anterior and posterior dorsal histoblast nests. Scale bar: 50 µM. Refers to Figure 5—figure supplements 4, 5.
miR-965 overexpression.
Early division arrest in a pupa overexpressing miR-965 under esg-GAL4 control (genotype: esg-GAL4, UAS-nuclear GFP/UAS-miR-965). Animals were collected for imaging at 0 hr APF. Scale bar: 50 µM. Refers to Figure 6C.





