TALPID3 controls centrosome and cell polarity and the human ortholog KIAA0586 is mutated in Joubert syndrome (JBTS23)
Figures
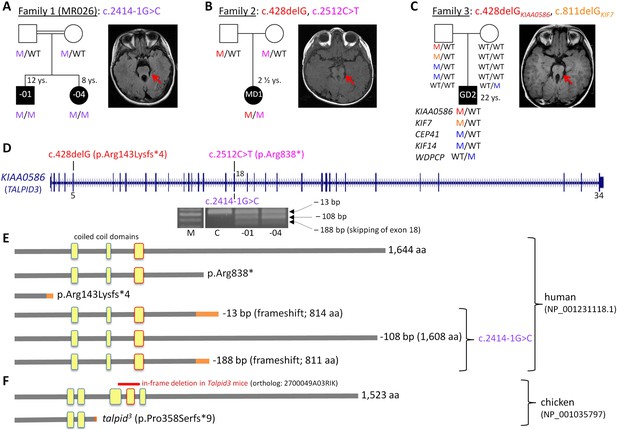
Patients with Joubert syndrome (JBTS) and KIAA0586 mutations (A–C).
(WT, wildtype; M, mutation). The ‘molar tooth sign’ in cranial axial MRI is indicated by arrows. (A) Family 1: Homozygosity mapping yielded eight homozygous chromosomal candidate regions (not shown), including the JBTS23 locus comprising KIAA0586. Patients MR026-01 and MR026-04 carry a homozygous splice site mutation, c.2414-1G>C. (B) Patient MD1 of Family 2 is compound heterozygous for two truncating mutations, including the prevalent c.428delG (p.Arg143Lysfs*4) allele. (C) Family 3: Patient G2 is double heterozygous for c.428delG in KIAA0586, and a frameshift mutation in KIF7 (JBTS12; c.811delG, p.Glu271Argfs*51). He also carries three potentially pathogenic variants in the ciliopathy genes CEP41, KIF14, and WDPCP (blue). (D) Genomic structure of KIAA0586 with mutations in exons 5 and in/adjacent to exon 18 indicated. The gel electrophoresis shows the aberrant transcripts due to c.2414-1G>C. (E) Scheme of human KIAA0586 protein and predicted consequences of JBTS-associated mutations. Orange color: unrelated residues included due to frameshift mutations. The third coiled-coil domain is the counterpart of the functionally essential fourth coiled-coil domain in chicken (framed in red). (F) Chicken TALPID3 (KIAA0586) is highly similar to the human protein. The talpid3 mutation results in an early frameshift and loss of three coiled-coil domains, including the fourth one. The in-frame deletion of exons 11 and 12 of mouse KIAA0586 (2700049A03Rik) is depicted above the scheme of the chicken ortholog.
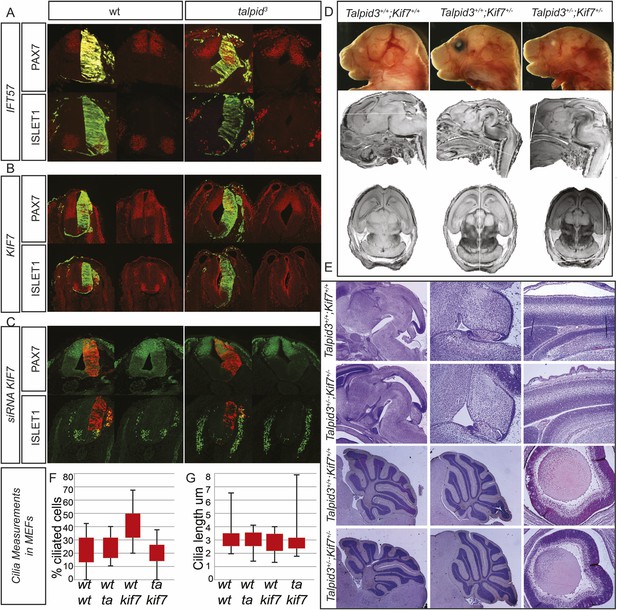
Analysis of potential interactions between Talpid3/TALPID3, Kif7/KIF7 and IFT57 in the mouse and in chicken.
Biallelic KIF7 mutations cause JBTS type 12 in human (Dafinger et al., 2011). Although both the KIAA0586 mutation c.428delG and the KIF7 mutation c.811delG were paternally inherited in patient G2, we sought to test for subtle abnormalities resulting from this double heterozygosity. In addition, we had previously found through a microarray analysis of talpid3 limb buds that IFT57, a protein associated with ciliopathy phenotypes in mice and zebrafish, is downregulated in talpid3 embryos (Bangs et al., 2011). Using in ovo complementation of the talpid3 neural tube, we could not detect induction of ISLET1 expression or ventralized PAX7 in the wildtype or talpid3 neural tube by overexpression of KIF7 or IFT57 (Figure 1—figure supplement 1A,B). We then used siRNA constructs against KIF7 to model a heterozygous loss of KIF7 in the TALPID3+/− neural tube. Knock-down with two siRNA constructs had a weak effect on neural tube patterning compared to the mouse KIF7−/− knockout (Cheung et al., 2009, Liem et al., 2009). Although the NKX2.2 expression domain could be marginally expanded in wildtype embryos (not shown), there was no expansion of ISLET1-positive motorneuron progenitors in wildtype or TALPID3+/− embryos. PAX7, however, was weakly dorsalized in both wildtype and talpid3+/− embryos (Figure 1FC). These results suggested that some KIF7 function may be cilia-independent as has been suggested (Liem et al., 2009). To more precisely investigate for a possible epistatic relationship between Kif7 and Talpid3, particularly in the organs primarily affected in JBTS, such as the cerebellum, we undertook a Talpid3+/− × Kif7+/− mouse cross in order to determine if double Talpid3+/−/Kif7+/− heterozygous animals had brain patterning malformations. We first dissected embryos at E15.5, 16.5, and 17.5 and found that Talpid3+/−/Kif7+/− embryos were morphologically normal, including size, situs and limb patterning. MEFs derived from E12.5 embryos were normally ciliated, with the percentage of ciliated cells and cilia length comparable to those seen in wildtype, Talpid3+/+/Kif7+/− and Talpid3+/−/Kif7+/+cells (Figure 1—figure supplement 1F,G). MRI and sectioning of the brain also showed no brain patterning abnormalities (Figure 1—figure supplement 1D,E). Subsequently, Talpid3+/−/Kif7+/− animals were born and grew normally compared to their litter mates and showed no abnormal brain morphology (Figure 1—figure supplement 1). We conclude that KIAA0586 (TALPID3) and KIF7 do not act epistatically and hypothesize that additional genetic alterations in ciliopathy genes of patient G2, eventually including those identified in CEP41 (JBTS15), KIF14, and WDPCP, may contribute to a mutational load that is sufficient to elicit a JBTS phenotype on a KIAA0586+/−; KIF7+/− background. (A) Overexpression of IFT57 does not have an effect on patterning of the neural tube in the talpid3 chicken. (B) Overexpression of KIF7 does not rescue or alter neural tube patterning in the talpid3 chicken. (C) siRNA knockdown of KIF7 resulted in a weak dorsalization of PAX7 but no expansion of ISLET1. (D) Talpid3+/− Kif7+/− mice showed no gross anatomical abnormalities, neither were developmental brain defects identified through MRI (D) or histology (E). (F, G) No abnormalities were identified in either the percentage of ciliated cells (F), nor the length of cilia (G) in MEFs derived from wildtype, Talpid3+/+Kif7+/−, Talpid3+/− Kif7+/+ or Talpid3+/− Kif7+/− mice.
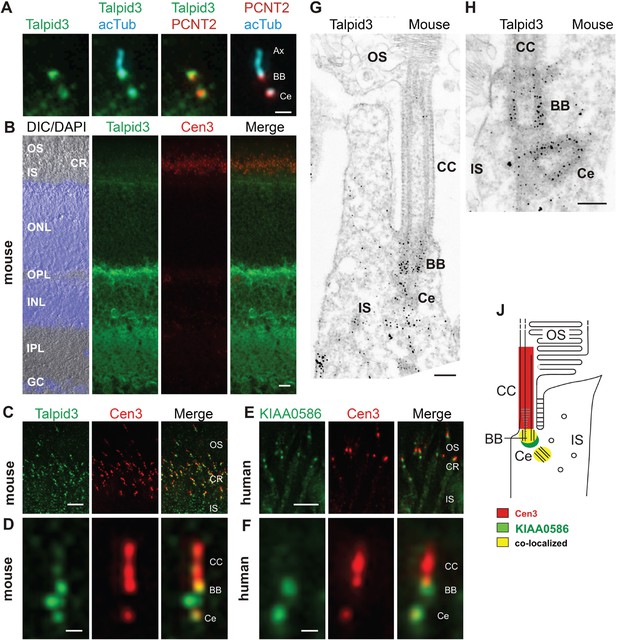
Localization of KIAA0586/Talpid3 in primary cilia and in photoreceptor cilia of mammalian retinas.
(A) Triple labeling of a ciliated IMCD3 cell demonstrates localization of Talpid3 (green) in the basal body (BB) and the adjacent centriole (Ce) at the base of the primary cilium co-stained by antibodies against Pericentrin-2 (PCNT2, red) and anti-acetylated tubulin (acTub, cyan), a biomarker of the axoneme (Ax). (B) Longitudinal cryosections through a mouse retina stained for Talpid3 (green) and counterstained for the ciliary marker Centrin-3 (Cen3, red) and for the nuclear DNA marker DAPI reveal Talpid3 localization in the ciliary region (CR) at the joint between the inner (IS) and the outer segment (OS) of the photoreceptor layer, the outer (OPL) and inner plexiform layer (IPL). Overlay of DIC (differential interference contrast) image with DAPI (blue) nuclear stain in the outer (ONL) and the inner nuclear layer (INL) and in the ganglion cell layer (GC). (C–F) Immunostaining of cryosections through the photoreceptor layer of a mouse (C) and a human retina (E) demonstrates co-localization of KIAA0586/Talpid3 and Cen3 in the CR of photoreceptor cells. Higher magnification of double-labeled mouse (D) and human (F) photoreceptor cilium reveals substantial localization of Talpid3/KIAA0586 at the centriole (Ce), the BB and between the Ce and BB of the photoreceptor cilium, but not in the connecting cilium (CC). (G, H) Immunoelectron microscopy analysis of longitudinal section through the cilium of a mouse rod photoreceptor cell and (G) higher magnification of the ciliary base (H) labeled for Talpid3 reveals Talpid3 in the periciliary region namely in the Ce and BB. (J) Schematic representation of Talpid3/KIAA0586 localization in the photoreceptor cilium. Scale bars: A, 1 μm; B, 10 μm; C, E, 5 μm; D, F, 0.5 µm; G, H, 200 nm.
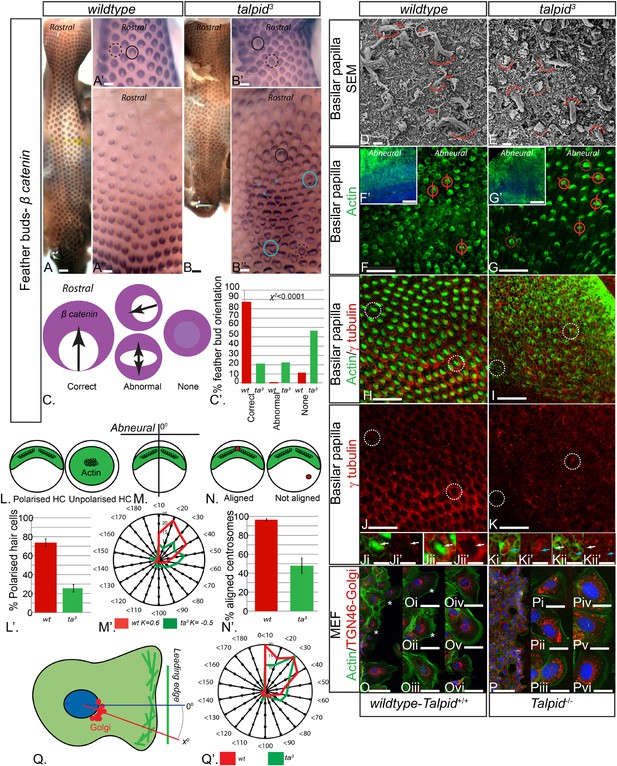
Loss of TALPID3 (KIAA0586) causes abnormal tissue and cell polarity and abnormal intracellular organization.
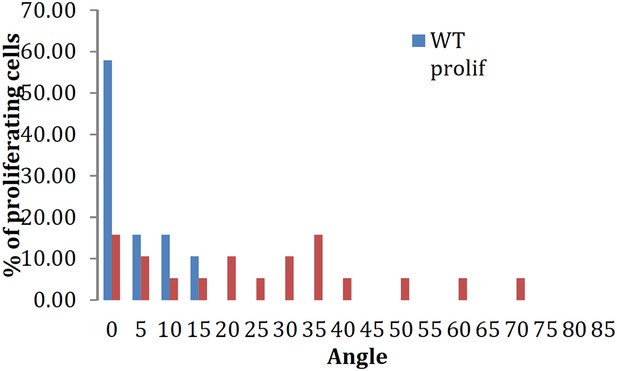
(A, A′, A′′) β-catenin expression is localized anteriorly within feather buds of the wildtype chicken at day 9.5 and (B, B′, B′′) in the talpid3 chicken at day 9.5. Black circles indicate feather buds with correct polarity; dashed black circles represent no polarity; blue circles represent abnormal polarity (Schematic C). The talpid3 chicken (B′′) demonstrates feather buds which lack polarity (blue circles) which is not seen in the wildtype chicken (A′′). Asterisks represent merged feather buds. (C′) Quantification of the percentage of feather buds with correct, abnormal or no polarity in wildtype and talpid3 (D, E) SEM of the basilar papilla in wildtype (D) and talpid3 (E) chickens. Arrows indicate cilia. Curved lines represent the base of stereocilia hair bundles. (F–K) Actin bundles identified by phalloidin (green) and centriolar localization identified by γ tubulin (red). (F′, G′) overview of wildtype and talpid3 basilar papilla, higher magnification in (F, G), red circles with line represent orientation of polarized actin bundles in basilar papilla; dashed red circles represent unpolarized actin bundles (Schematic L, M). (L′) Quantification of polarized haircells. (M′) Quantification of the angle of polarised hair cells. (H–K) Dashed white circles represent magnified images (Ji–Kii′). (Ji–Kii) White arrows indicate aligned centrosomes; blue arrows indicate unaligned centrosomes (Schematic N). (N′) Quantification of cells with aligned centrosomes. (O, P) Orientation based on placement of Golgi (TGN46, red) in comparison to actin indicating the leading edge (phalloidin, green) and nucleus (Dapi, blue, schematic in Q) in MEFs. Asterisks represent areas of higher magnification (not all represented at lower magnification). (Q′) Quantification of the angle of orientation of MEF cells in scratch assay. Scale Bars: A, B 5 mm; A′, A′′, B′, B′′ 1 mm; D, E 1 μm; F, G, H, I, J, K 20 μm; F′, G′ 100 nm; Ji, Ji′, Jii, Jii′, Ki, Ki′, Kii, Kii′ 10 μm; O, P 100 μm; Oi, Oii, Oiii, Oiv, Ov, Ovi, Pi, Pii, Piii, Piv, Pv, Pvi 25 μm.
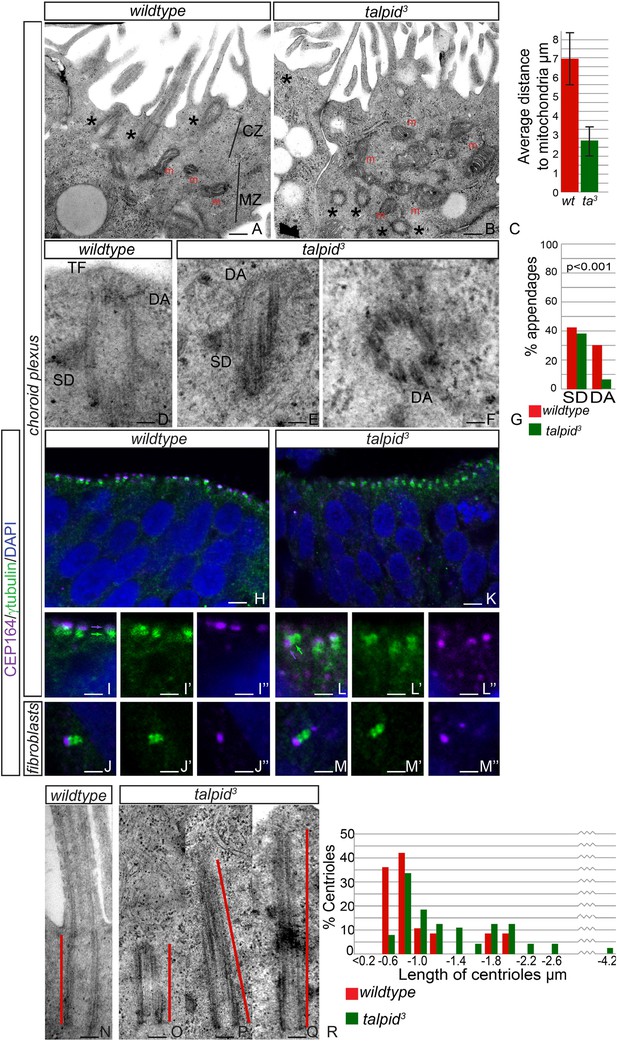
Loss of TALPID3 causes abnormal intracellular organization and centriolar orientation
(A, B) The chicken choroid plexus at E8 is a highly polarized structure with docked centrioles (asterisk, A) identified within a clear centriolar zone apically (CZ, A) and a mitochondrial zone (MZ; m indicates mitochondria). The talpid3 choroid plexus (B) lacks these defined zones, with mitochondria identified in the most apical zone (m, B) centrioles identified throughout the cell, failing to dock (asterisk, B). Quantification of distance of mitochondria to cell surface (C). (D–G) talpid3 tissue is capable of producing mature centrioles. Wildtype centrioles (D) and talpid3 centrioles (E, F) exhibited subdistal appendages (SD), and distal appendages (DA), although DA were less frequently observed on talpid3 centrioles, quantified in (G). CEP164 localizes to the distal mother centriole in wildtype and talpid3 choroid plexus neuroepithelium (purple arrow indicated distal mother centriole, green arrow proximal centriole; H, I, I′, I′′, K, L, L′, L′′) and fibroblasts (J, J′, J′′, M, M′, M′′), but CEP164 puncta are smaller and disorganized in talpid3 choroid plexus and fail to orientate to the apical surface of the cell (arrows L). Centrioles in wildtype tissue were on average 0.7 µm (red line indicating centriole/basal body; N, R) compared to 0.9 µm in the talpid3 choroid plexus (O, P, Q, R). Scale bars: A, B = 1 μm, D, E, F = 100 nm; H, K = 10 μm I, J, L, M = 5 μm, N, O, P, Q = 200 nm.
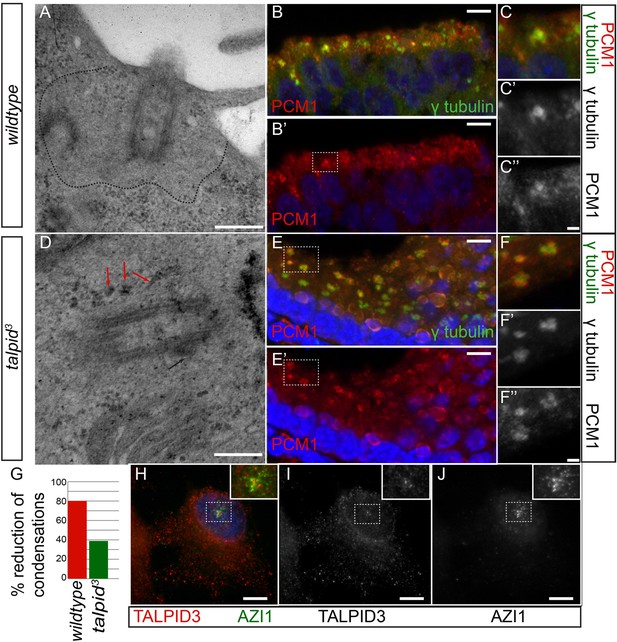
Analysis of centriolar satellites in the talpid3 choroid plexus.
An area clear of electron-dense condensations was observed around the basal body in wildtype cells (area outlined by dots; A), electron-dense condensations were observed adjacent to talpid3 centrioles (indicated by arrows, D). Quantified in (G). Immunostaining for a centriolar satellite marker in the choroid plexus, PCM1 (magnified area outlined by dashed line; PCM1 = red, γ tubulin, green B, B′, C, C′, C′′, E, E′, F, F′, F′′). KIAA0586 protein does not colocalize with AZI1, a satellite protein in human RPE1 cells (KIAA0586 = red, AZI1 = green H, I, J). Scale bars: A, D = 500 nm; B, E 10 = μm; C, F = 2 μm H, I, J 5 μm.