RNA binding protein Caprin-2 is a pivotal regulator of the central osmotic defense response
Figures
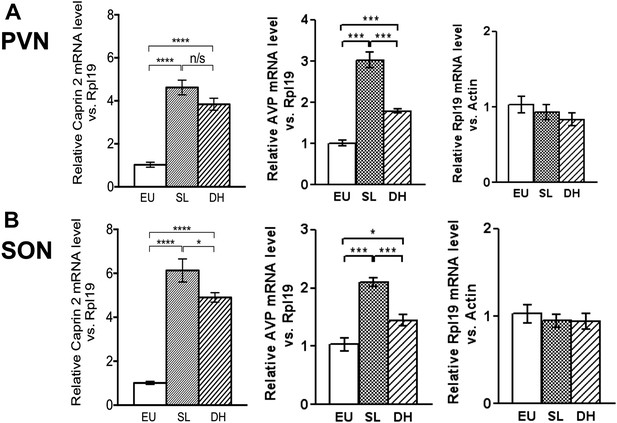
Caprin-2 messenger ribonucleic acid (mRNA) expression in the rat paraventricular nuclei (PVN) and supraoptic nuclei (SON) increases following a chronic osmotic stimulus.
Quantitative RT-PCR analysis (qRT-PCR) analysis of Caprin-2 mRNA expression in the PVN (A) and SON (B) of euhydrated (EU), salt-loaded (SL) and dehydrated (DH) rats. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001, n = 5, One-way ANOVA with Sidak's post-hoc test. Compared with euhydrated (EU) rats, both SL and DH resulted in significant up-regulation of Caprin-2 mRNA in the PVN and SON. SL and DH significantly increased Caprin-2 mRNA levels in the PVN (euhydrated PVN, 1.03 ± 0.04; salt-loaded PVN, 4.62 ± 0.34; dehydrated PVN, 3.84 ± 0.28; n = 5; p ≤ 0.0001 salt-loaded and dehydrated vs euhydrated). In the SON, SL further increased the Caprin-2 mRNA levels compared to DH (p < 0.05) and SON (euhydrated SON, 1.01 ± 0.07; salt-loaded SON, 6.13 ± 0.53; dehydrated SON, 4.9 ± 0.22 p ≤ 0.0001 salt-loaded and dehydrated vs euhydrated, salt-loaded vs dehydrated). In parallel, arginine vasopressin (AVP) mRNA levels are significantly increased in both PVN (euhydrated PVN, 1.01 ± 0.07; salt-loaded PVN, 3.03 ± 0.18; dehydrated PVN, 1.79 ± 0.05; n = 6; p ≤ 0.001 salt-loaded and dehydrated vs euhydrated, salt-loaded vs dehydrated) and SON (euhydrated SON, 1.03 ± 0.11; salt-loaded, SON 2.10 ± 0.07; dehydrated SON, 1.45 ± 0.09 p ≤ 0.001 salt-loaded vs euhydrated, p ≤ 0.05 dehydrated vs euhydrated, and p ≤ 0.001 salt-loaded vs dehydrated). Rpl19 levels are unchanged in both the PVN (euhydrated PVN, 1.03 ± 0.11; salt-loaded PVN, 0.93 ± 0.10; dehydrated PVN, 0.83 ± 0.09, ns) and SON (euhydrated SON, 1.03 ± 0.10; salt-loaded SON, 0.95 ± 0.08; dehydrated SON 0.94 ± 0.09, ns).
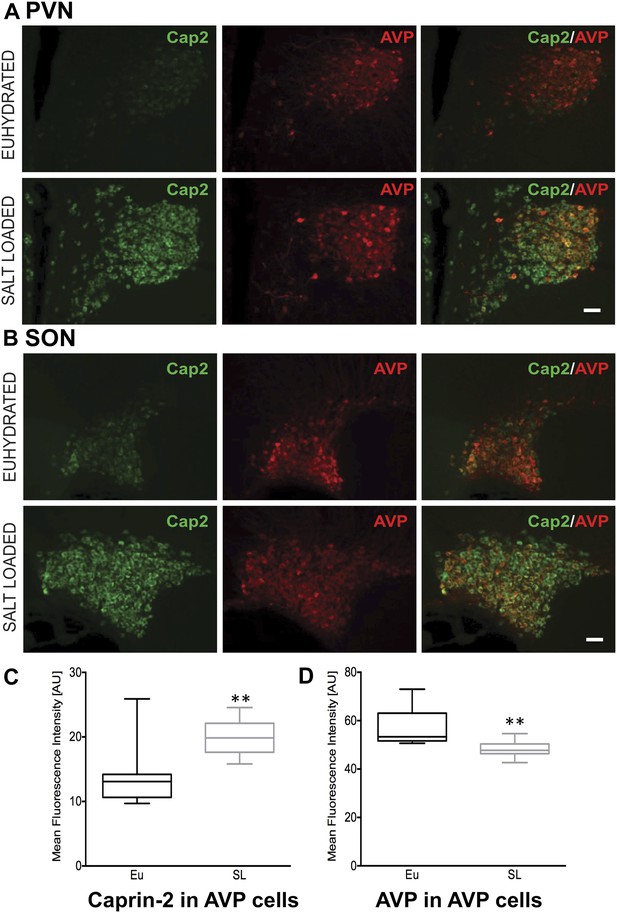
Caprin-2 protein expression in the rat PVN and SON.
Immunohistochemical analysis of Caprin-2 protein expression (green) in the rat PVN (A) and SON (B) in euhydrated and salt-loaded rats; co-localization with AVP-neurophysin II (AVP; red). Scale bar 100 µm. (C) Quantification of fluorescence signals for Caprin-2 and (D) AVP in AVP cells. Mean fluorescence intensities were quantified in 10 SON wide field fluorescence microscope images acquired from 3 rats. AVP signal was selected (above the same threshold for EU and SL samples) and mean fluorescence intensity was measured for Caprin 2 and AVP signal. Each result was corrected by subtraction of mean background intensities for each channel. Salt-loading (SL) significantly increases Caprin-2 signal in AVP cells (euhydrated, 13.68 ± 1.47; salt-loaded, 19.92 ± 0.86, p = 0.0018), but results in a significant decrease of AVP in AVP cells (euhydrated, 56.94 ± 2.439; salt-loaded 48.3 ± 1.05, p = 0.0044). **p ≤ 0.01.
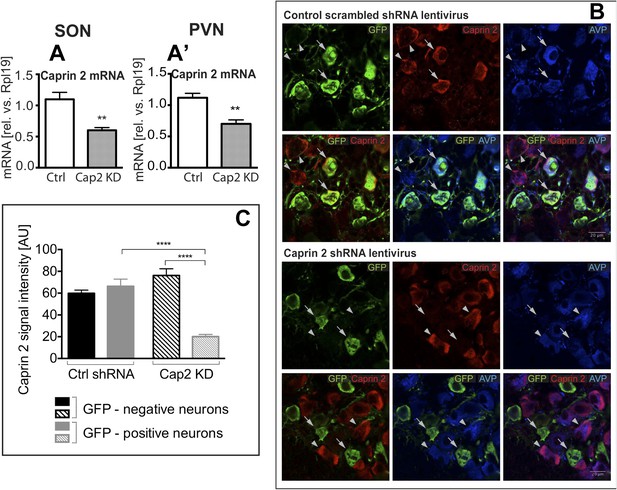
Lentivirus-mediated Caprin-2 shRNA knockdown in the SON and PVN.
qRT-PCR analysis of the effect of Caprin-2 knockdown in the SON (A) and PVN (A′) on Caprin-2 mRNA levels (1.10 ± 0.11 vs 0.60 ± 0.04 in the Ctrl, n = 18, and Cap2 KD, n = 11, SON, p = 0.0023; 1.03 ± 0.07 vs 0.64 ± 0.06 in the Ctrl, n = 16, and Cap2 KD, n = 7, PVN, p = 0.0017). GAPDH mRNA levels are unchanged by Cap2 KD (1.03 ± 0.07 vs 1.04 ± 0.1, in the Ctrl, n = 18, and Cap2 KD, n = 11, SON, n.s.; 1.02 ± 0.05 vs 1.01 ± 0.1 in the Ctrl, n = 16, and Cap2 KD, n = 7, PVN, n.s.). (B, C) Immunohistochemistry-based quantification of Caprin-2 gene knockdown in magnocellular neurones (MCNs). (B) MCNs in the SON transduced with control scrambled shRNA and Caprin-2 shRNA lentiviruses visualized by immunostaining for eGFP (GFP, green) and co-localised with Caprin-2 (red) and AVP neurophysin II (AVP; blue). Full arrows—GFP positive cells. Arrow heads—GFP negative cells. Whilst the scrambled shRNA has no effect on Caprin-2 levels, in cells expressing the specific Caprin-2 shRNA, Caprin-2 expression is much reduced. (C) There was no significant difference in Caprin-2 signal between eGFP-negative and positive neurons in the Ctrl rats (respectively, 59.86 ± 2.89 and 66.25 ± 6.61; n = 18 and 23; n.s.) In contrast, in Cap2 KD SON, Caprin-2 signal was significantly reduced in eGFP-positive neurons compared to eGFP-negative cells (respectively, 20.16 ± 1.97 vs 76.19 ± 6.22; n = 15 and 26; p ≤ 0.0001). **p ≤ 0.01, ****p ≤ 0.0001.
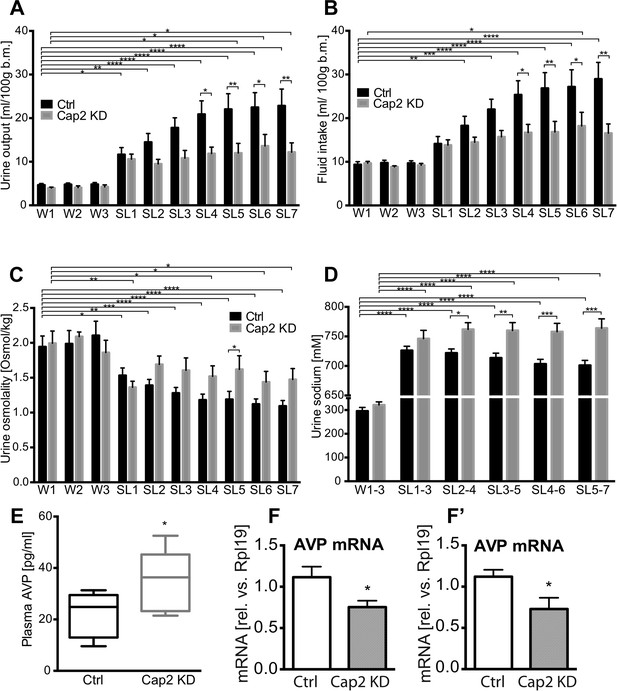
Physiological effects of Caprin-2 gene knockdown in euhydrated and salt-loaded rats.
Urine output (A), fluid intake (B), urine osmolality (C) and urine sodium concentration (D) were measured in control, scrambled shRNA (Ctrl) and Caprin-2 shRNA lentivirus-injected (Cap2 KD) euhydrated (received water for 3 days, W1–3) and salt-loaded rats (received 2% wt/vol NaCl ad libitum for 7 days, SL1-7). Plasma AVP concentration (E) was measured after 7 days of SL, at the end of the experiment. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001, n = 9 (Ctrl) and 5 (Cap 2 KD). (F, F′) qRT-PCR analysis of the effects of Caprin-2 knockdown in the SON (F) and PVN (F′) on AVP mRNA levels (1.12 ± 0.13 vs 0.75 ± 0.08 for Ctrl, n = 18, and Cap2 KD, n = 11, SON, p = 0.047; 1.06 ± 0.08 vs 0.68 ± 0.13 for Ctrl, n = 16, and Cap2 KD, n = 7, PVN, p = 0.019). *p ≤ 0.05.
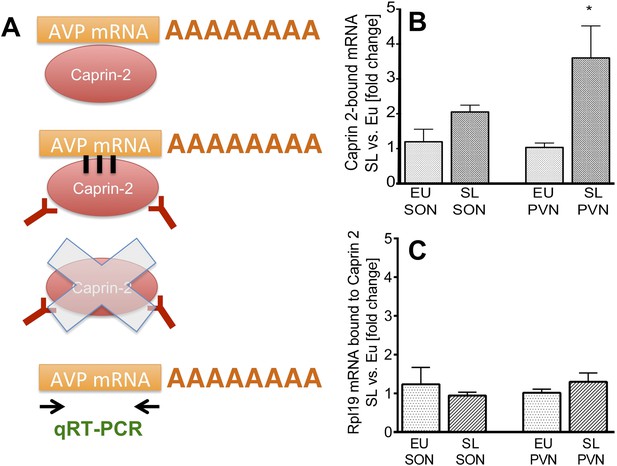
Caprin-2 binds to the AVP mRNA in the SON and PVN.
Binding of AVP by Caprin-2 protein in the SON and PVN of euhydrated (EU) and salt-loaded (SL) rats determined by RNA immunoprecipitation assay. (A) In the RNA immunoprecipitation assay, SON or PVN tissue punches from EU or SL rats were first exposed to formaldehyde in order to covalently cross-link RNA with associated proteins. Cell extracts were then incubated with antibodies recognizing Caprin-2. Following immunoprecipitation, and hence enrichment of specific complexes, cross-links were reversed and extracted RNA was subject to qRT-PCR to detect AVP mRNA sequences. (B) Effects of salt-loading on the amount of Caprin-2 binding to AVP mRNA in the SON (1.2 ± 0.36 EU vs 2.05 ± 0.20 SL, n = 5, p = 0.072) and PVN (1.03 ± 0.13 EU vs 3.6 ± 0.92; n = 5, p = 0.0243). (C) Salt-loading has no effect on the amount of Caprin-2 binding to Rpl19 mRNA in the SON and PVN. *p ≤ 0.05.
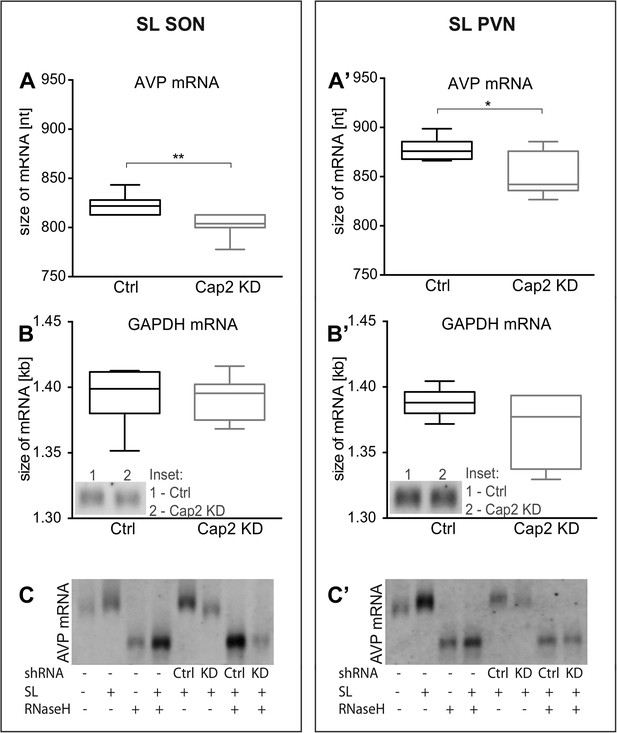
In vivo effects of Caprin-2 gene knockdown on the length of the AVP mRNA poly(A) tail.
AVP mRNAs were analyzed by Northern Blotting. Quantification results of AVP (A, A′) and GAPDH mRNA (B, B′) in samples obtained from the SON (A, B) and PVN (A′, B′) of SL rats injected with lentivirus with either, scrambled shRNA (Ctrl) or Caprin-2 shRNA (Cap2 KD). Insets on the plots with GAPDH mRNA represent typical images. Representative images of AVP mRNA before and after removing poly(A) tails (RNaseH) in the SON (C) and PVN (C′) of euhydrated (−) and salt-loaded (SL) rats, as well as in salt-loaded rats injected with scrambled (Scr shRNA) or Caprin-2 (Cap2 shRNA) shRNA. Quantification of transcript sizes revealed that Caprin-2 knockdown prevented the SL-induced increase of the AVP mRNA length, reducing it from 822.7 ± 3.76 nt. (n = 8, Ctrl) to 801.7 ± 4.47 nt. (n = 7, Cap2 KD) (p = 0.0031) in the SON, and from 877.7 ± 4.77 nt. (n = 6, Ctrl) to 851.3 ± 9.23 nt. (n = 6, Cap2 KD) in the PVN (p = 0.0296). No size changes were observed with the GAPDH mRNA (Ctrl SON, n = 8, 1393 ± 7.55 nt. vs Cap2 KD, n = 7, 1392 ± 6.12 nt., n.s.; Ctrl PVN, n = 6, 1388 ± 4.65 nt. vs Cap2 KD, n = 6, 1368 ± 11.24 nt., n = 6, n.s.). *p ≤ 0.05, **p ≤ 0.01.
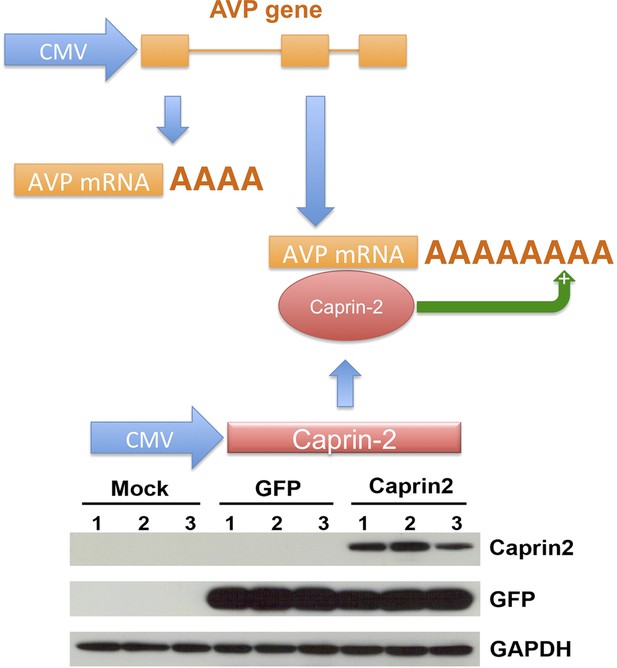
A recapitulated in vitro system for examining effect of Caprin-2 on the metabolism of the AVP mRNA.
In order to further examine to role of Caprin-2 in AVP mRNA metabolism, we developed a recapitulated in vitro system. We overexpressed the rat AVP structural gene and full-length rat Caprin-2 in HEK293T cells, both under the control of the heterologous CMV promoter. A vector expressing eGFP was used as a control. Western blotting of triplicate independent samples showing robust expression of eGFP and Caprin-2 proteins in transduced cells.
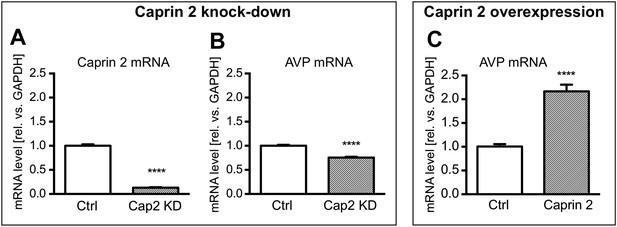
Effects of Caprin-2 overexpression or knockdown on AVP mRNA levels in vitro.
For Caprin-2 knockdown experiments, cells were co-transfected with the CMV-driven rat Caprin-2 overexpression construct, the CMV-driven rat AVP structural gene, and either scrambled shRNA (Ctrl) or Caprin-2 shRNA (Cap2 KD) constructs. Caprin-2 knockdown (Cap2 KD) reduces (A) Caprin-2 (Ctrl, 1.00 ± 0.03 vs Cap2 KD, 0.13 ± 0.01, n = 5, p ≤ 0.0001) and (B) AVP (Ctrl, 1.00 ± 0.02 vs Cap2 KD, 0.76 ± 0.02; n = 5, p ≤ 0.0001) mRNA levels. In the Caprin-2 overexpression experiments, the CMV-driven rat AVP structural gene was co-transfected with either the CMV-driven rat-Caprin-2 overexpression construct (Cap2), or a control CMV-eGFP construct (Ctrl). (C) Caprin-2 overexpression increases the level of AVP mRNAs (Ctrl, 1.0 ± 0.05 vs Cap2, 2.17 ± 0.14, n = 5, p ≤ 0.0001). ****p ≤ 0.0001.
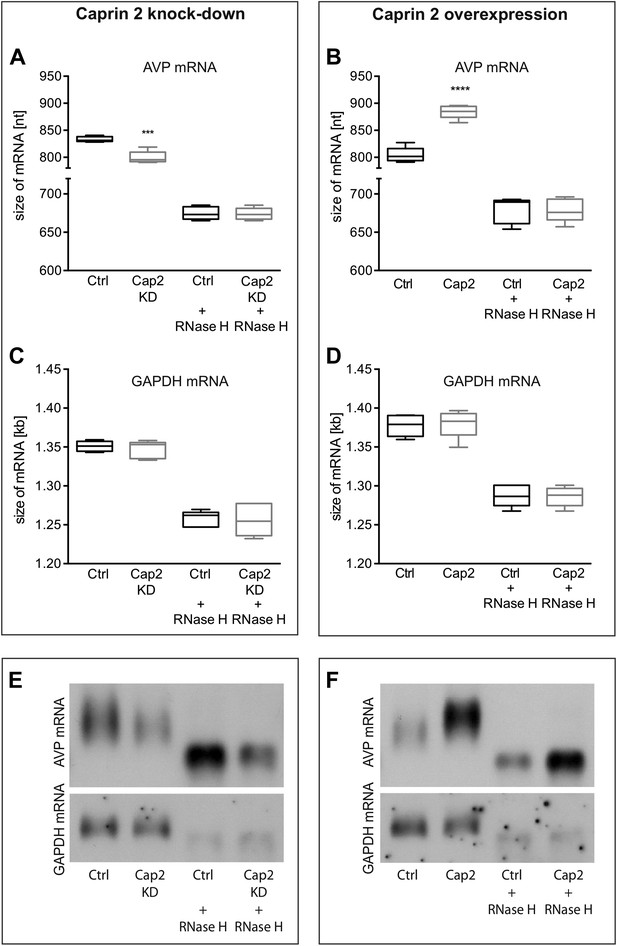
Effects of Caprin-2 overexpression or knockdown on AVP mRNA poly(A) tail length in vitro.
Effects of Caprin-2 knockdown (Cap2 KD) (A) and overexpression (Cap2) (B) in HEK293 cells on the length of the poly(A) tail in AVP mRNA, analyzed by Northern Blotting. Knockdown reduces (Ctrl, 833.3 ± 2.21 nt. vs Cap2 KD, 799.6 ± 5.07 nt., n = 5, p = 0.0003) whilst overexpression increases (Ctrl, 804.4 ± 6.19 nt. vs Caprin-2, 884.3 ± 5.51 nt., n = 5; p ≤ 0.0001) the length of the AVP mRNA. This is due to modulation in the lenth of the poly(A) tail, when this is removed there are no differences between the groups (Ctrl, 674.8 ± 3.75 nt. vs Cap2 KD, 674 ± 3.48 nt., n = 5; n.s.; Ctrl, 679 ± 7.62 nt. vs Cap2, 678.9 ± 6.79 nt., n = 5; n.s.). Neither Caprin-2 knockdown (C) (Ctrl, 1351 ± 2.94 nt., vs Cap2 KD, 1347 ± 4.98 nt., n = 5, n.s.) nor Caprin-2 overexpression (D) (Ctrl, 1377 ± 6.13 nt. vs Caprin-2, 1380 ± 8.05 nt., n = 5; n.s.) had any effect on the size of the GAPDH mRNA. Typical images representing the effects of Caprin-2 knockdown (E) and overexpression (F) on AVP mRNA in HEK293T cells before and after removing poly(A) tails (RNaseH) are shown. ****p ≤ 0.0001, ***p ≤ 0.0003.
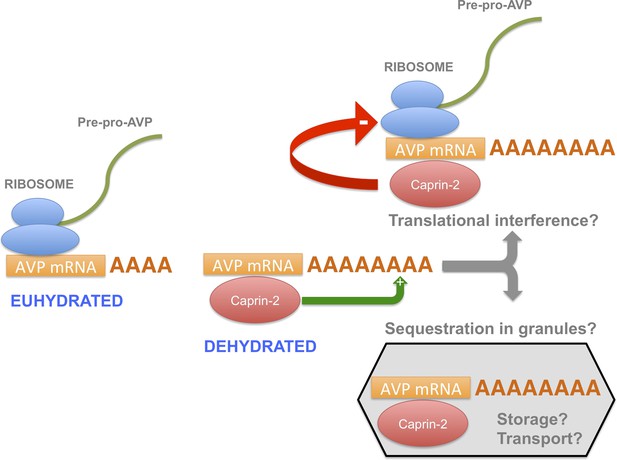
A model for the actions of Caprin-2.
Caprin-2 expression increases in the rat SON and PVN following an osmotic stimulus. Caprin-2 binds to the AVP mRNA, and mediates an increase in the length of the poly(A) tail, either by stimulating further synthesis, or by inhibiting deadenylation. Caprin-2 also mediates an increase in VP mRNA abundance, possibly by increasing transcript stability. Paradoxically, we propose that Caprin-2 inhibits translation of the AVP mRNA, perhaps by displacing ribosomes or by slowing initiation or elongation, or through mediating sequestration into translationally inert storage or transport granules.
Tables
Urine output (A), fluid intake (B), Urine osmolality (C) and urine sodium concentration (D) in rats injected into the SON and PVN with either, control, scrambled shRNA or Caprin-2 shRNA, in euhydrated (water: W1–3) and salt-loading (SL 1–7) conditions
| Scrambled shRNA | Caprin 2 shRNA | |||||
|---|---|---|---|---|---|---|
| Mean | SEM | N | Mean | SEM | N | |
| A. Urine output (ml/100 g b.m.) | ||||||
| W1 | 4.661 | 0.344 | 9 | 3.954 | 0.222 | 5 |
| W2 | 4.784 | 0.339 | 9 | 4.048 | 0.423 | 5 |
| W3 | 4.803 | 0.413 | 9 | 4.230 | 0.461 | 5 |
| SL1 | 11.649 | 1.590 | 9 | 10.600 | 1.155 | 5 |
| SL2 | 14.482 | 2.005 | 9 | 9.488 | 1.062 | 5 |
| SL3 | 17.790 | 2.294 | 9 | 10.814 | 1.776 | 5 |
| SL4 | 20.884 | 3.086 | 9 | 11.860 | 1.485 | 5 |
| SL5 | 22.046 | 3.564 | 9 | 11.982 | 2.223 | 5 |
| SL6 | 22.473 | 3.367 | 9 | 13.598 | 2.666 | 5 |
| SL7 | 22.833 | 3.828 | 9 | 12.194 | 2.135 | 5 |
| B. Fluid intake (ml/100 g b.m.) | ||||||
| W1 | 9.353 | 0.668 | 9 | 9.674 | 0.421 | 5 |
| W2 | 9.752 | 0.607 | 9 | 8.832 | 0.251 | 5 |
| W3 | 9.684 | 0.542 | 9 | 9.203 | 0.425 | 5 |
| SL1 | 14.127 | 1.665 | 9 | 13.806 | 1.224 | 5 |
| SL2 | 18.270 | 2.145 | 9 | 14.483 | 1.156 | 5 |
| SL3 | 22.002 | 2.325 | 9 | 15.707 | 1.460 | 5 |
| SL4 | 25.351 | 3.211 | 9 | 16.680 | 1.886 | 5 |
| SL5 | 26.842 | 3.580 | 9 | 16.818 | 2.437 | 5 |
| SL6 | 27.166 | 3.912 | 9 | 18.209 | 3.124 | 5 |
| SL7 | 28.952 | 3.805 | 9 | 16.554 | 2.108 | 5 |
| C. Urine osmolality (mOsmol/kg) | ||||||
| W1 | 1943.333 | 154.937 | 9 | 1994.000 | 175.516 | 5 |
| W2 | 1986.667 | 190.343 | 9 | 2090.000 | 65.651 | 5 |
| W3 | 2105.556 | 204.702 | 9 | 1860.000 | 176.437 | 5 |
| SL1 | 1532.222 | 108.189 | 9 | 1362.000 | 87.772 | 5 |
| SL2 | 1390.000 | 85.261 | 9 | 1690.000 | 120.831 | 5 |
| SL3 | 1280.000 | 80.035 | 9 | 1606.000 | 178.342 | 5 |
| SL4 | 1181.111 | 83.773 | 9 | 1520.000 | 149.767 | 5 |
| SL5 | 1188.889 | 115.548 | 9 | 1620.000 | 195.090 | 5 |
| SL6 | 1121.111 | 75.064 | 9 | 1438.000 | 152.918 | 5 |
| SL7 | 1093.333 | 79.861 | 9 | 1474.000 | 156.831 | 5 |
| D. Urine sodium (mM) | ||||||
| W1-3 | 295.929 | 15.179 | 25 | 321.306 | 13.196 | 15 |
| SL1-3 | 726.221 | 7.102 | 27 | 746.221 | 14.048 | 15 |
| SL2-4 | 721.850 | 6.852 | 27 | 761.959 | 11.233 | 15 |
| SL3-5 | 713.653 | 7.925 | 27 | 759.992 | 13.389 | 15 |
| SL4-6 | 703.270 | 7.888 | 27 | 758.025 | 14.093 | 15 |
| SL5-7 | 701.085 | 8.319 | 27 | 763.926 | 15.838 | 15 |
Additional files
-
Supplementary file 1
The rat Caprin-2 gene. (a) The sequence of full-length rat brain Caprin-2 cDNA. (b) The predicted amino acid sequence of full-length rat brain Caprin-2 protein. (c) Alternatively spliced isoforms of rat brain Caprin-2. (d) Hypothetical functional domains of the rat brain Caprin-2 protein based on alignment to the Xenopus RNG105 protein sequence, a paralogue of the well-analyzed rat Caprin-1 protein, which is highly homologous to Caprin-2.
- https://doi.org/10.7554/eLife.09656.014





