Non-classical amine recognition evolved in a large clade of olfactory receptors
Figures
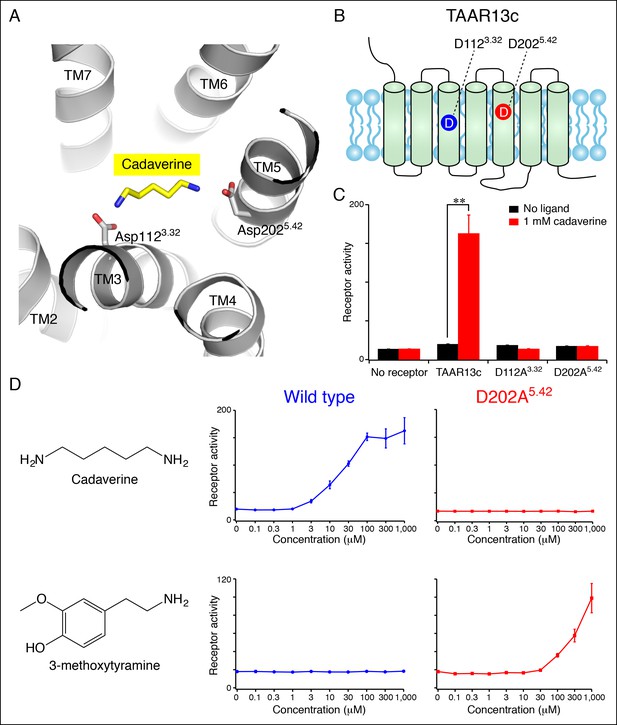
Diamine recognition sites in the cadaverine receptor TAAR13c.
(A) Structural modeling of zebrafish TAAR13c bound to cadaverine (yellow) reveals two aspartates (D1123.32 and D2025.42) with carboxylates (red) predicted to form salt bridges to ligand amino groups (blue). (B) Cartoon of zebrafish TAAR13c topology depicts the location of D1123.32 and D2025.42. (C) Charge-neutralizing mutation of D1123.32 and D2025.42 abrogates cadaverine responsiveness of zebrafish TAAR13c expressed in HEK-293 cells (mean ± sem, n = 3, **p<0.01 based on two-tailed unpaired Student's t-test). (D) D202A5.42 mutation transforms TAAR13c from a diamine receptor into a monoamine receptor (mean ± sem, n = 3).
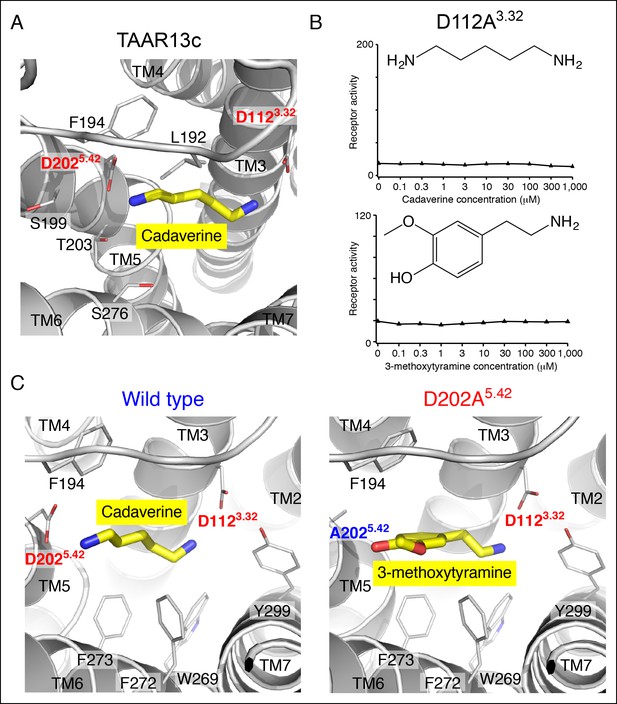
Functional analysis and structural modeling of TAAR13c mutants.
(A) Structural modeling of TAAR13c bound to cadaverine (yellow) reveals residues within 5 Å of the amino group pointing towards transmembrane α-helix 5. (B) Charge-neutralizing mutation of Asp3.32 eliminates cadaverine responsiveness in TAAR13c. HEK-293 cells were co-transfected with Cre-Seap and the D112A3.32 mutant of TAAR13c, incubated with cadaverine or 3-methoxytyramine at various concentrations, and assayed for phosphatase activity (mean ± sem, n = 3). (C) Structural modeling of wild type TAAR13c bound to cadaverine (yellow) and the TAAR13c D202A5.42 mutant bound to 3-methoxytyramine (yellow).
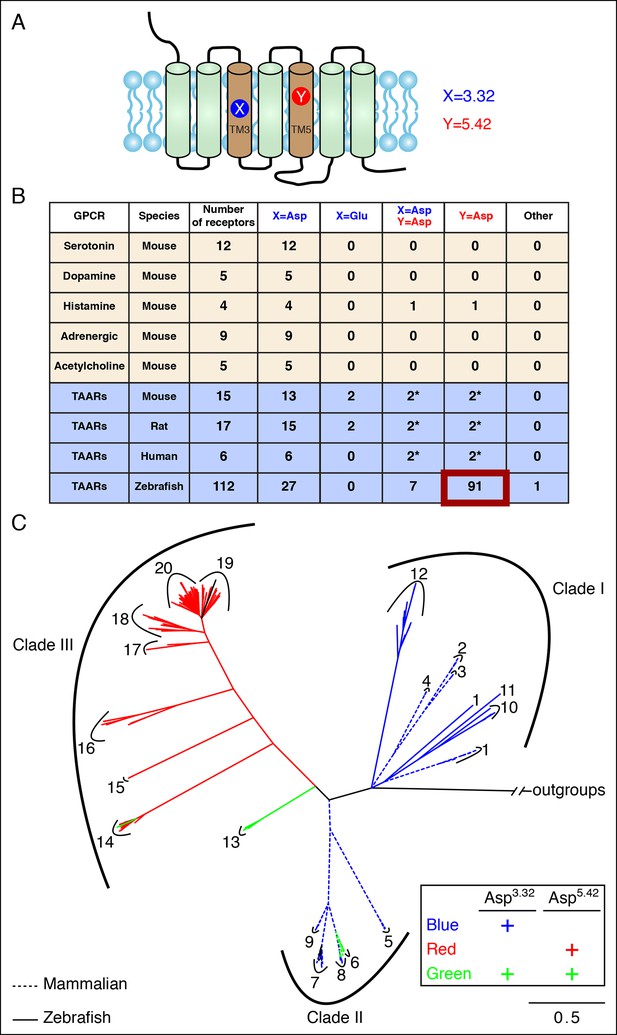
Asp5.42 is highly conserved in clade III TAARs.
(A) Cartoon depiction indicates the location of positions 3.32 and 5.42 in GPCRs. (B) Bioinformatic analysis reveals the number of receptors with anions at positions 3.32 (X) or 5.42 (Y) across various biogenic receptor subfamilies in mouse, as well as TAARs in mouse, rat, humans, and zebrafish. Two mouse, rat, and human TAARs have Asp5.43 instead of Asp5.42, and these are included in columns marked Y = Asp (*). (C) Phylogenetic analysis of the TAAR family in zebrafish (solid lines), mice (dashed lines), rats (dashed lines), and humans (dashed lines); scale bar = 0.5 substitutions per site. TAARs containing only Asp3.32 (blue), only Asp5.42 (red), or both Asp3.32 and Asp5.42 (green) are depicted. Mammalian TAARs with Asp3.32 and Asp5.43 instead of Asp5.42 are green. Glu3.32-containing rodent TAARs and the one zebrafish TAAR, TAAR19f, that lacks both Asp3.32 and Asp5.42 are depicted in black.
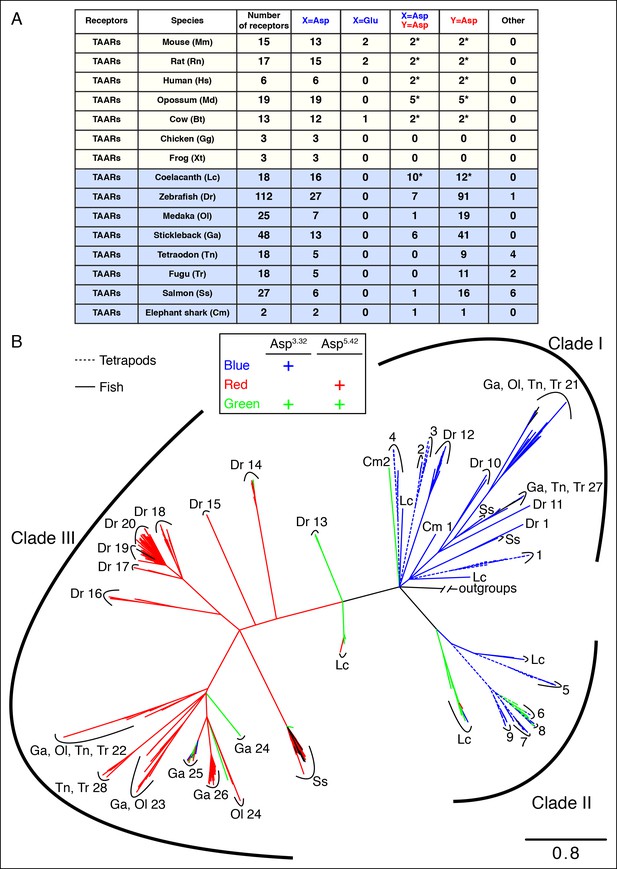
The phylogenetic tree of tetrapod and fish TAARs.
(A) As in Figure 2, bioinformatic analysis reveals the number of receptors with anions at positions 3.32 (X) or 5.42 (Y). (B) Phylogenetic analysis of the TAAR family in fish (solid lines) and tetrapods (dashed lines); scale bar = 0.8 substitutions per site. TAARs containing only Asp3.32 (blue), only Asp5.42 (red), both Asp3.32 and Asp5.42 (green), and neither (black) are depicted.
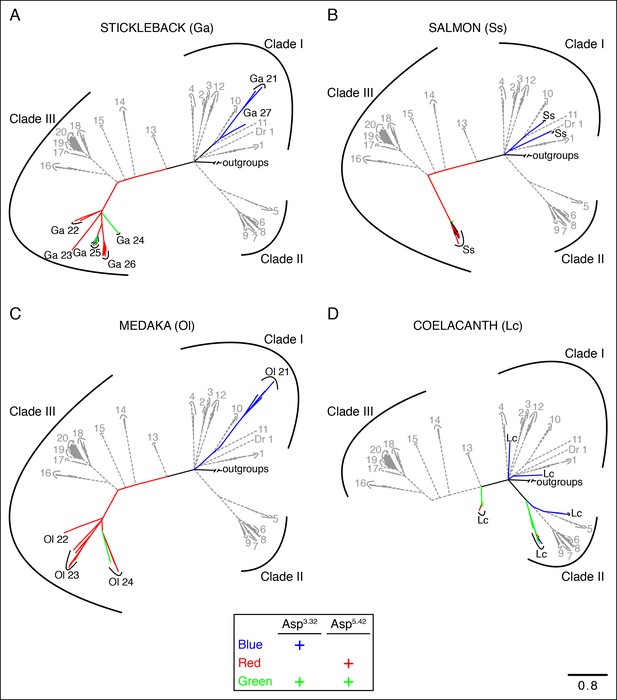
Phylogenetic analysis of TAARs from different fish species.
Using the phylogenetic tree in Figure 2—figure supplement 1B as a template, TAARs are highlighted from individual fish species indicated (solid, color), as well as from zebrafish and tetrapods (dashed, grey); scale bar = 0.8 substitutions per site. TAARs containing only Asp3.32 (blue), only Asp5.42 (red), both Asp3.32 and Asp5.42 (green), or neither (black) are depicted.
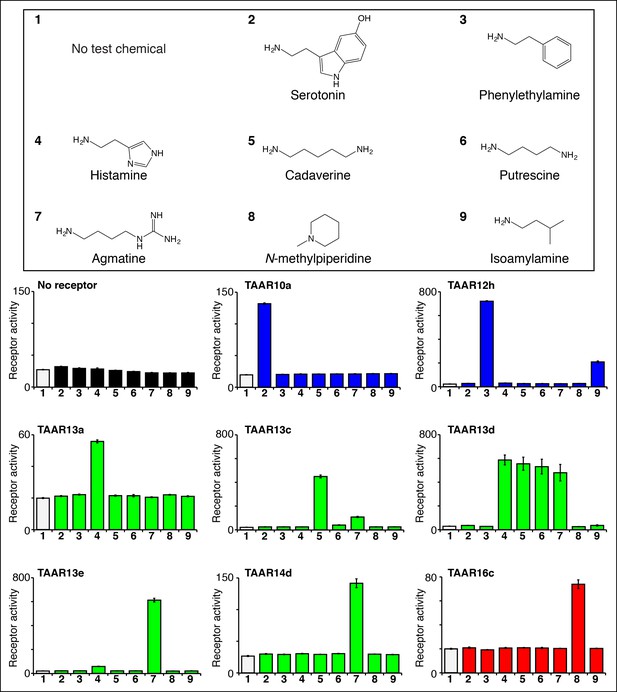
Identifying the first ligands for several zebrafish TAARs.
HEK-293 cells were cotransfected with Cre-Seap and plasmids encoding zebrafish TAARs, incubated with test chemicals (100 μM), and assayed for phosphatase activity using a fluorescent substrate (mean ± sem, n = 3). Zebrafish TAAR10a and TAAR12h are clade I TAARs containing Asp3.32 but not Asp5.42 (blue), zebrafish TAAR13a, TAAR13c, TAAR13d, and TAAR13e, and TAAR14d contain both Asp5.42 and Asp3.32 (green), and zebrafish TAAR16c is a clade III TAAR containing Asp5.42 but not Asp3.32 (red).
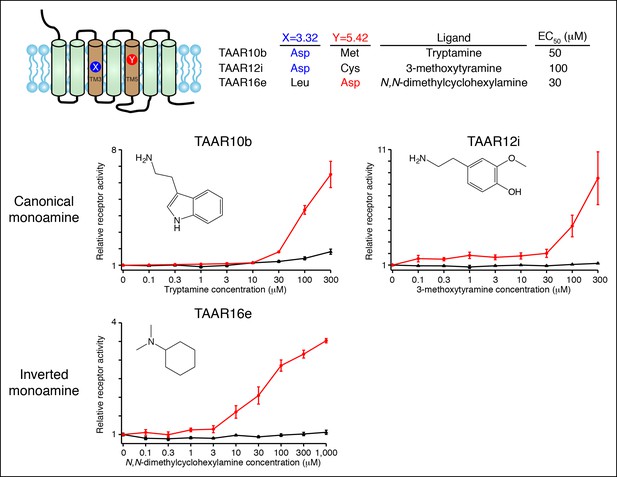
Functional expression of TAAR10b, TAAR12i, and TAAR16e in Hana3A cells.
Hana3A cells were co-transfected with Cre-Seap and TAAR-encoding plasmids, incubated with ligands, and assayed for phosphatase activity (mean ± sem, n = 6).
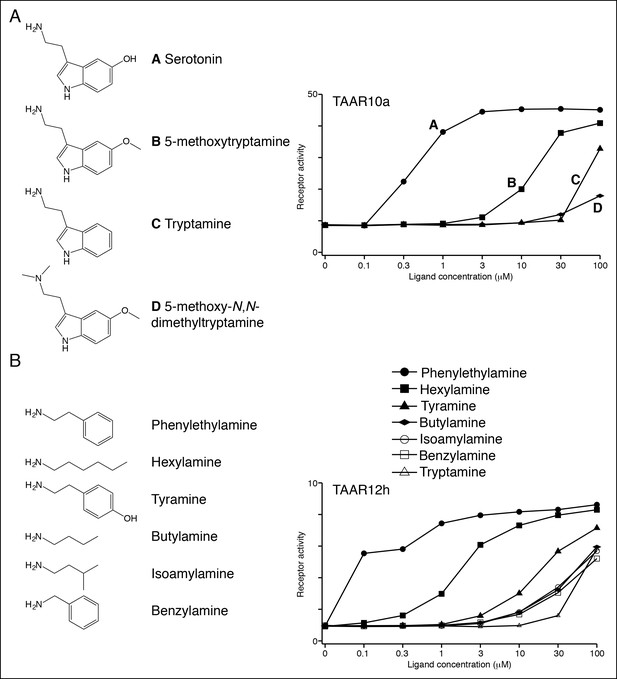
Structure-function studies of zebrafish clade I TAARs: TAAR10a and TAAR12h.
HEK-293 cells were co-transfected with Cre-Seap and TAAR-encoding plasmids, incubated with ligands, and assayed for phosphatase activity.
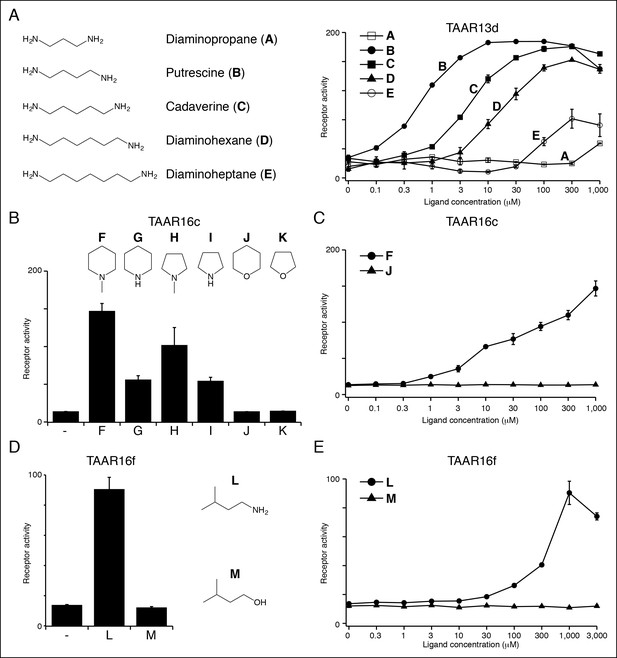
Structure-activity studies of zebrafish TAARs.
(A) Zebrafish TAAR13d displays highest affinity for putrescine among tested ligands, and reduced affinity for longer or shorter diamines. (B) Zebrafish TAAR16c recognizes several structurally related amines but not oxygen-containing analogs (1 mM). F: N-methylpiperidine; G: piperidine; H: N-methylpyrrolidine; I: pyrrolidine; J: tetrahydropyran; K: tetrahydrofuran. (C) Dose-dependent responses of TAAR16c for N-methylpiperidine and tetrahydropyran. (D) Zebrafish TAAR16f recognizes isoamylamine (L) but not isoamyl alcohol (M) at 1 mM. (E) Dose-dependent responses of TAAR16f for isoamylamine and isoamyl alcohol (mean ± sem, n = 3).
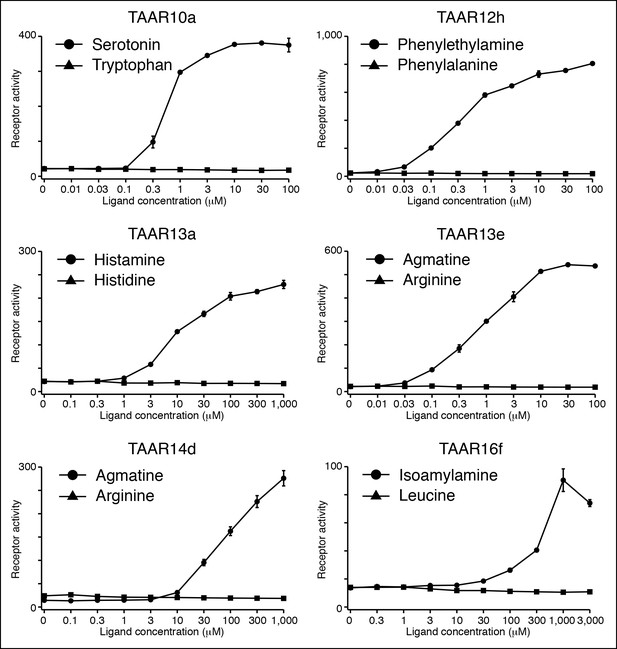
Several TAARs detect amines but not amino acids.
HEK-293 cells were co-transfected with Cre-Seap and TAAR-encoding plasmids, incubated with ligands, and assayed for phosphatase activity (mean ± sem, n = 3).
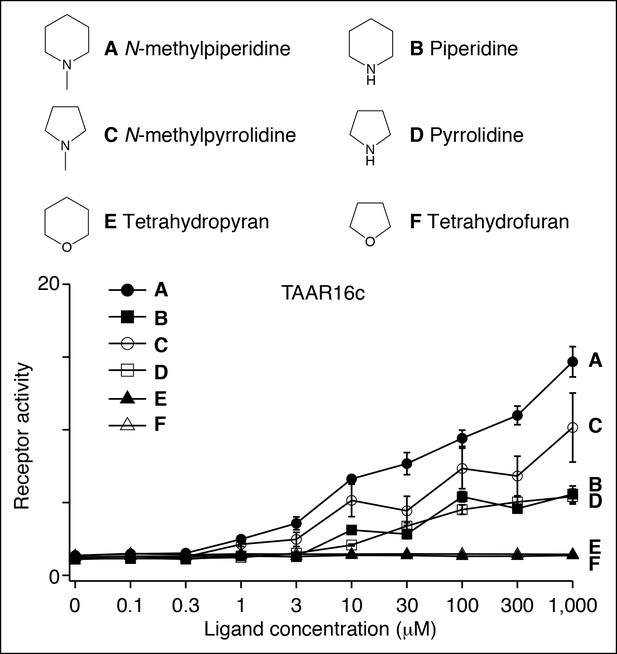
Structure-function studies of the zebrafish clade III TAAR, TAAR16c.
HEK-293 cells were co-transfected with Cre-Seap and plasmid encoding TAAR16c, incubated with ligands, and assayed for phosphatase activity (mean ± sem, n = 3–6).
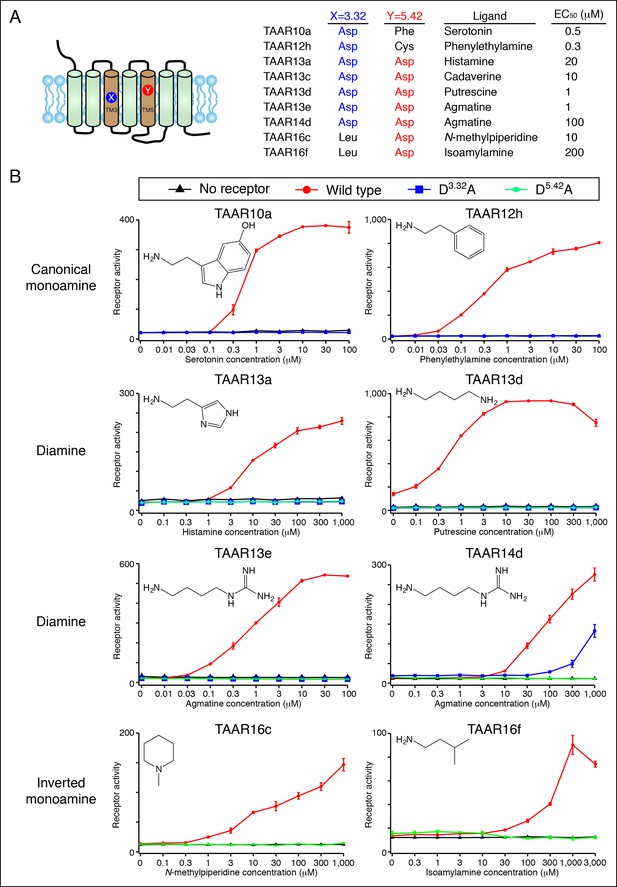
Dose-dependent responses of zebrafish TAARs and charge-neutralizing TAAR mutants.
(A) The identities of amino acids 3.32 and 5.42 in nine 'de-orphaned' zebrafish TAARs, as well as preferred ligands and corresponding EC50s, are depicted. (B) Dose-dependent activation of TAARs and TAAR mutants by ligands indicated (mean ± sem, n = 3). Responses are shown in cells transfected with Cre-Seap alone (black) or together with wild type TAARs (red), D3.32A mutant TAARs (blue), and D5.42A mutant TAARs (green). D3.32 is lacking in clade III TAARs (TAAR16c, TAAR16f) and D5.42 is lacking in clade I TAARs (TAAR10a, TAAR12h), so the corresponding D3.32A and D5.42A mutants could not be generated.
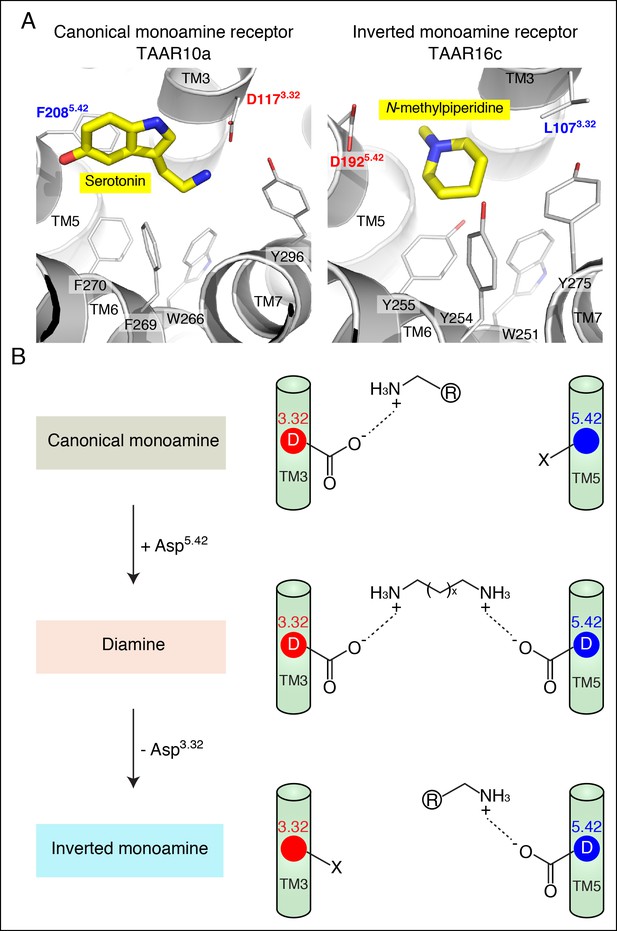
Modeling the structure and evolution of non-classical amine recognition.
(A) Structural modeling of zebrafish TAAR10a and TAAR16c bound to serotonin and N-methylpiperidine (yellow) respectively. (B) A model for the birth of a large clade of olfactory receptors with non-classical amine recognition. We propose that clade III TAARs evolved a non-classical mode of amine recognition in two steps. First, an ancestral TAAR gained the ability to recognize diamines by acquiring Asp5.42. Subsequently, a diamine-detecting TAAR lost the canonical amine recognition site, Asp3.32, leading to non-classical amine recognition through a transmembrane α-helix V salt bridge. Extensive gene duplication and mutation expanded and diversified clade III TAARs, leading to a large clade of olfactory receptors with non-canonical amine-detection properties.
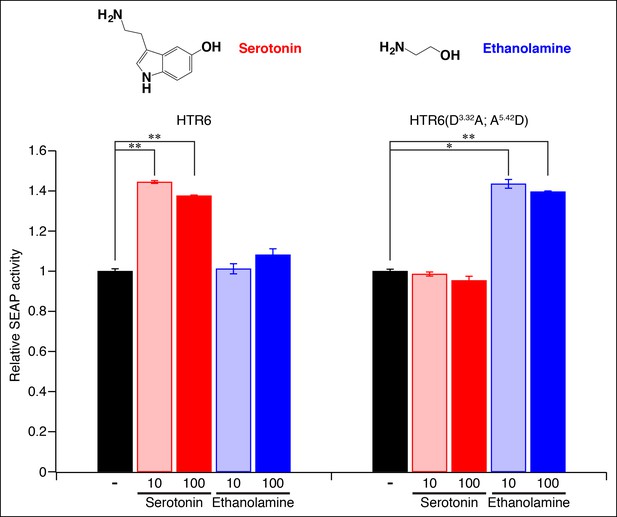
Re-engineering the amine contact site of HTR6.
HEK-293 cells were co-transfected with Cre-Seap and plasmids encoding HTR6 or an HTR6 double mutant (D3.32A; A5.42D), incubated with ligands (0, 10 or 100 μM), and assayed for phosphatase activity (mean ± sem, n = 3, *p <. 05, **p <. 01 based on one-way ANOVA analysis and Dunnett’s test to compare controls and ligand-activated responses).





