Precise regulation of the guidance receptor DMA-1 by KPC-1/Furin instructs dendritic branching decisions
Figures
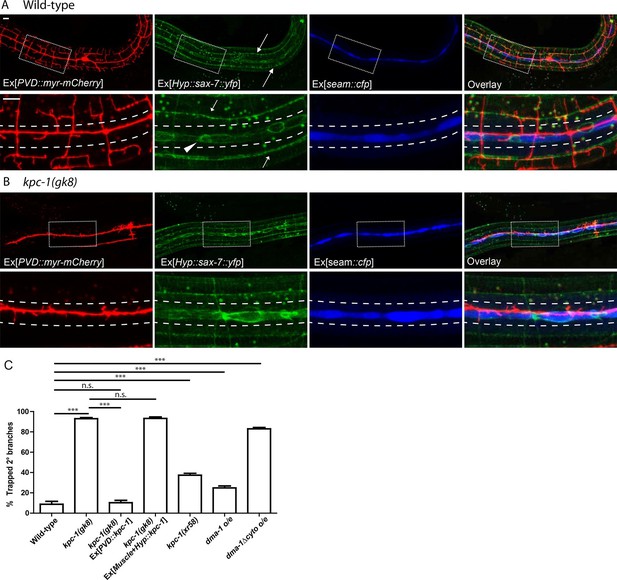
The kpc-1 mutants showed severe trapping of PVD dendrites.
(A) Fluorescent images showing (red) morphology of the PVD neuron, (green) localization of SAX-7 in the hypodermal cell, (blue) seam cells and overlay between the three in wild-type worms. SAX-7 was enriched in two sublateral longitudinal lines and at the lateral midline around the seam cell-hypodermal junctions. Arrows: Sublateral stripes of enriched SAX-7 that co-localize with PVD 3° dendrites. Arrowhead: SAX-7 enriched near the 1°dendrites, where it was encountered by the 2° branches as they emerge. The images in the lower panels are zoomed-in views of the regions indicated by the boxes. Dotted lines indicate the 'trap zone' marked by enriched SAX-7 around seam cells. (B) In kpc-1(gk8) mutants, almost all branches failed to grow out of the trap zone between the dotted lines indicated by enriched SAX-7. Scale bar: 10 μm. (C) Quantification of the percentage of 2° branches trapped around the 1°dendrite. *** is p<0.001, n.s. is p>0.05 by Student’s T-test. N=50 for each genotype.
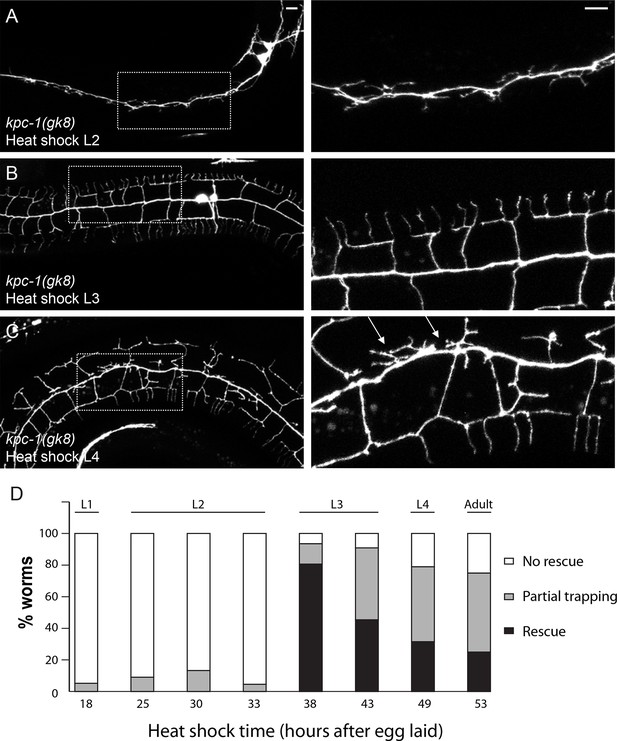
Heat shock expression of KPC-1 at distinct time points during development produced different phenotypes.
(A) Expressing KPC-1 during L2 failed to rescue the kpc-1(gk8) mutant phenotype. (B) Expressing KPC-1 during early L3 completely rescued the mutant phenotype. (C) Expressing KPC-1 during L4 or later stages could partially rescue some menorahs, but still many branches were trapped in the trap zone. Scale bar: 10 μm. (D) Quantification of the percentage of worms heat shocked at various time points showing no rescue, partial or full rescue.
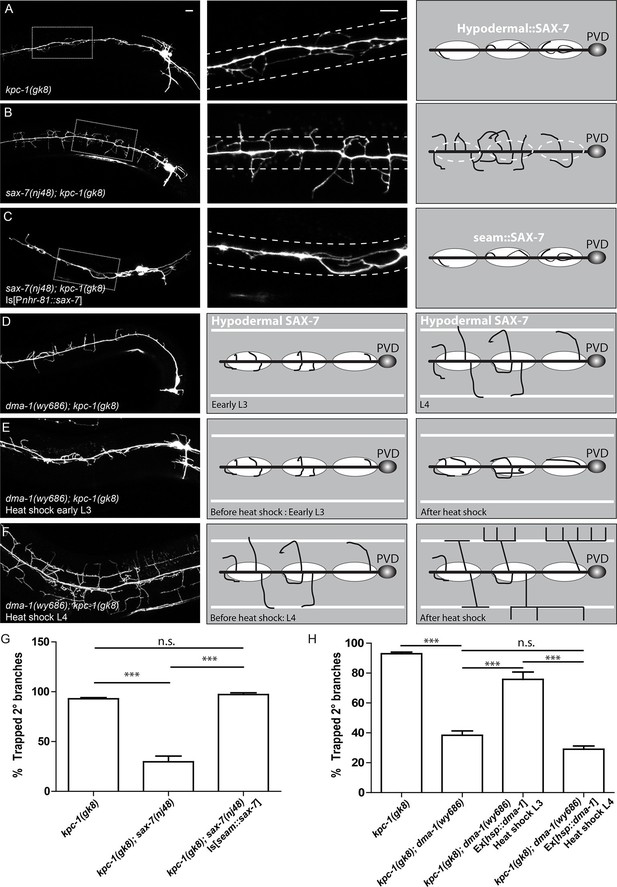
The SAX-7/MNR-1/DMA-1 tripartite complex was causal for the trapping phenotype in kpc-1 mutants.
(A) Left: Fluorescent image showing PVD morphology of a kpc-1(gk8) null mutant. Middle: Zoomed-in view of the boxed area in the left panel. Dotted lines indicate the 'trapping zone' with enriched SAX-7. Almost all 2° dendrites were trapped in this region. Right: Schematic illustration of the phenotype. (B) PVD morphologies of sax-7(nj48); kpc-1(gk8) double mutants were indistinguishable from sax-7(nj48) single mutants. Dendrites could escape from the trap zone. (C) Expressing SAX-7 in the seam cells restored the trapping phenotype. (D) Left: Fluorescent image showing the PVD morphology of a dma-1(wy686); kpc-1(gk8) double mutant. Middle: Schematic illustration showing the initial phase of 2° branch outgrowth during early L3 when the dendrites pass the 'trap zone'. Right: Later in development, dendrites of the dma-1; kpc-1 mutants had escaped the trap zone but failed to form menorahs due to lack of DMA-1. (E) Expressing DMA-1 during early L3 in dma-1(wy686); kpc-1(gk8) double mutants completely restored the trapping phenotype. (F) Expressing DMA-1 during L4 or later stages generated a striking rescue of menorahs. Since the dendrites had already escaped from the trap zone, supplying DMA-1 enabled the dendrites to respond to sublateral SAX-7 and MNR-1 signal and form normal 3° and 4° branches at the right place. Scale bar: 10 μm. (G-H) Quantification of the percentage of 2° branches that were trapped around the 1°dendrite. *** is p<0.001, n.s. is p>0.05 by Student’s T-test. N=50 for each genotype.
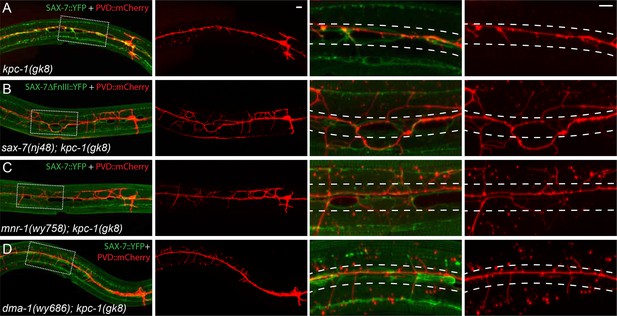
The SAX-7/MNR-1/DMA-1 ligand-receptor complex was causal for the trapping phenotype in kpc-1 mutants.
(A–D) Fluorescent images showing hypodermal SAX-7 localization in green and PVD morphology in red of (A) kpc-1(gk8) single mutant, (B) sax-7(nj48); kpc-1(gk8), (C) mnr-1(wy758); kpc-1(gk8) and (D) dma-1(wy686); kpc-1(gk8) double mutants. In the kpc-1 single mutant, almost all 2° branches failed to grow out of the trap zone indicated by the enriched SAX-7 (between dotted lines), whereas in double mutants, despite strong morphology defects, 2° dendrites grew out of the trap zone and extended toward the sublateral lines. Full length SAX-7::YFP was used in panels A, C and D while SAX-7ΔFnIII::YFP, a non-functional form of SAX-7 which still showed correct subcellular localization, was used in panel B to prevent rescuing of the sax-7 mutant phenotype. The images in the right 2 columns are zoomed-in views of the regions indicated by the dashed boxes on the left.
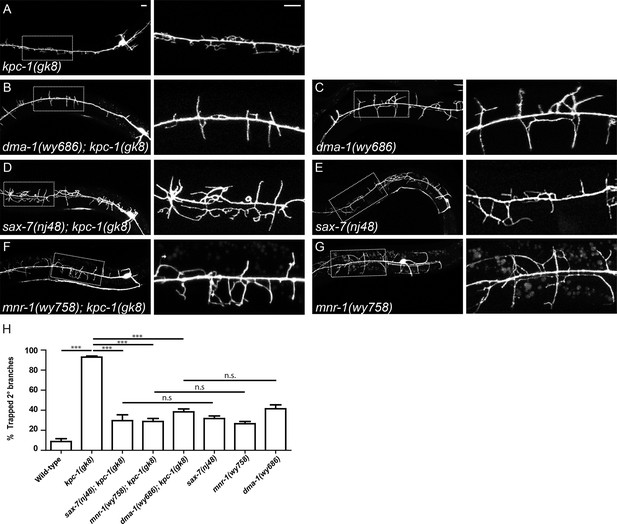
SAX-7, MNR-1 and DMA-1 were epistatic to KPC-1.
(A–G) Fluorescent images showing PVD morphologies of (A) kpc-1(gk8), (B) dma-1(wy686); kpc-1(gk8), (C) dma-1(wy686), (D) sax-7(nj48); kpc-1(gk8), (E) sax-7(nj48), (F) mnr-1(wy758); kpc-1(gk8) and (G) mnr-1(wy758) mutants. The PVD dendritic phenotypes of the double mutants were indistinguishable from the sax-7, mnr-1 and dma-1 single mutants but different from the kpc-1 single mutants. Scale bar: 10 μm. (H) Quantification of the percentage of 2° branches that were trapped around the 1°dendrite. *** is p<0.001, n.s. is p>0.05 by Student’s T-test. N=50 for each genotype.
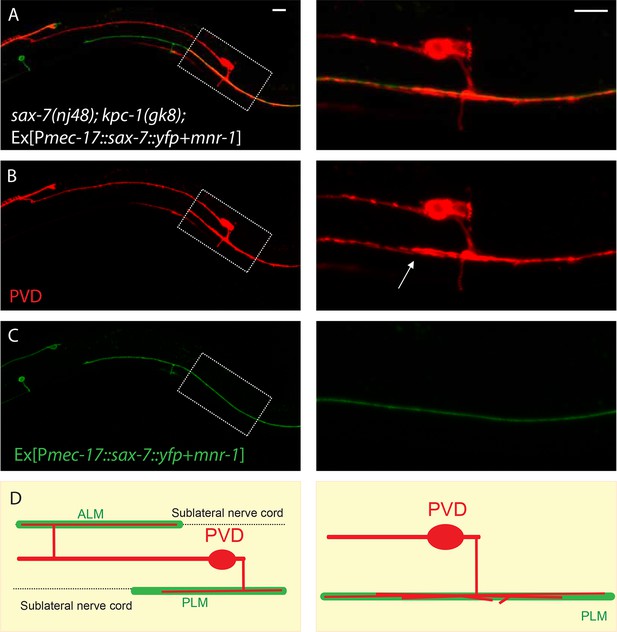
PLM and ALM neurons expressing SAX-7-YFP and MNR-1 caused the PVD dendrites of sax-7; kpc-1 double mutants to follow these neurons.
(A–C) Fluorescent images of (A) overlay, (B) PVD neuron and (C) PLM and ALM neurons expressing SAX-7::YFP and MNR-1. Right panels show the zoomed-in views of boxed regions in the left panels. Arrow points to multiple PVD dendrites fasciculating on the PLM neurite. Scale bar: 10 μm (D) Schematic illustration of the phenotype.
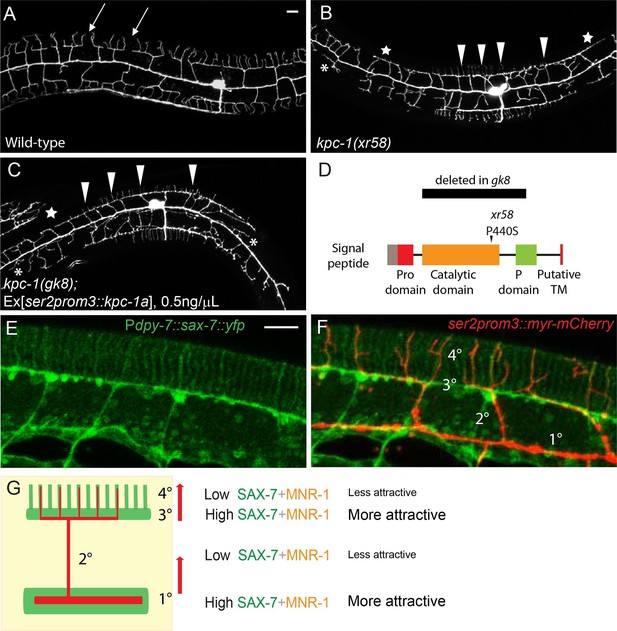
Partial loss of KPC-1 caused defects in higher order dendritic branches.
(A–C) Fluorescent images showing PVD morphologies of (A) wild-type (B) kpc-1(xr58) mutant and (C) kpc-1(gk8) mutant animals expressing a low concentration (0.5ng/μL) of full length, functional KPC-1. The xr58 mutants had severe defects in self-avoidance of 3° branches and reduced number of 4° branches. The phenotype was mimicked by expressing low-level wild-type KPC-1 in the gk8 null allele of kpc-1. Arrows: Gaps between 3° branches in wild-type neurons. Arrowheads: 3° branches that overlapped with their neighbors in mutants. Star: Defective menorah with no 4° branches. Asterisks: Trapped 2° branches. (D) Schematic of the KPC-1 protein showing locations of mutations in gk8 and xr58 mutants. (E–F) Fluorescent images of (E) SAX-7 localization in the hypodermal cell, and (F) overlay with a PVD marker in red. SAX-7 was highly enriched in the sublateral lines where 3° branches formed and grew and was also localized to vertical stripes followed by the 4° branches but at much lower concentration. Scale bar: 10 μm. (G) Schematic figure of PVD outgrowth. 2° branches emerging from the 1° dendrites and 4° branches growing away from the 3° branches faced similar challenges to go from regions with higher levels of SAX-7 to places that were less attractive. Compromised function of KPC-1 led to defects in escaping.
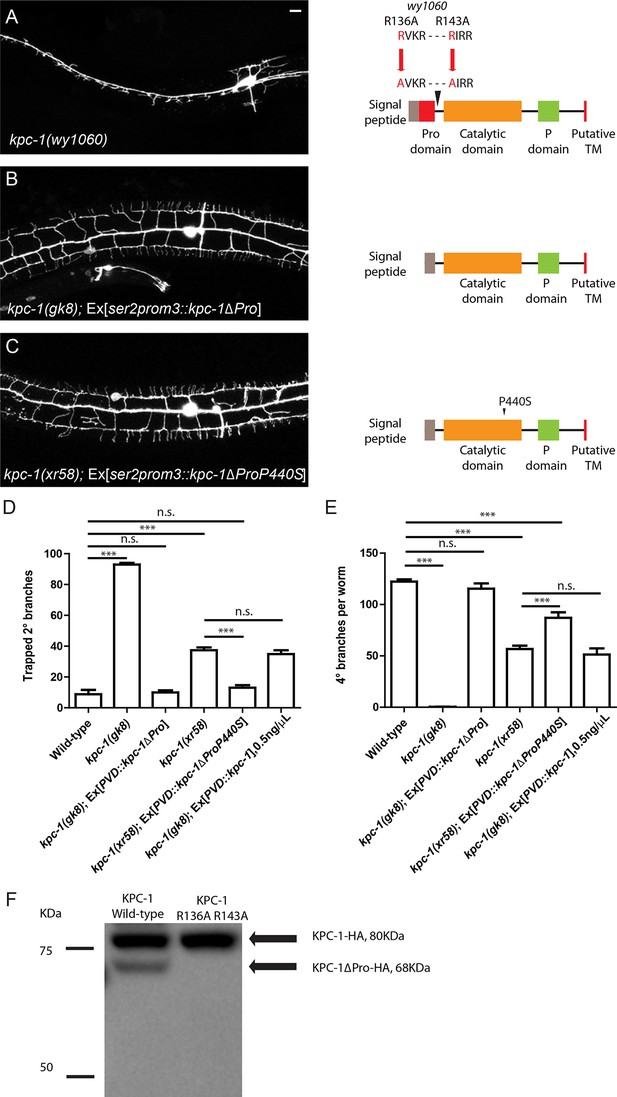
Activation of KPC-1 required self-cleavage.
(A) Fluorescent image showing PVD morphology of kpc-1(wy1060) mutants generated by CRISPR. Both self-cleavage sites were mutated (R136A, R143A). Schematic figure on the right shows the locations of the mutated self-cleavage sites. (B) KPC-1 lacking the N-terminal Pro domain could fully rescue the null kpc -1(gk8) mutants. Scale bar: 10 μm. (C) KPC-1ΔProP440S lacking its Pro domain and also carrying the P440S point mutation in the protease domain rescued the kpc-1(xr58) mutant phenotype. (D) Quantification of the percentage of 2° branches that were trapped around the 1°dendrite. *** is p<0.001, n.s. is p>0.05 by Student’s T-test. N=50 for each genotype. (E) Quantification of the total number of 4° branches per animal. *** is p<0.001, n.s. is p>0.05 by Student’s T-test. N=50 for each genotype. (F) Western blot showing HA-tagged KPC-1 expressed in S2 cells. Two bands were detected corresponding to full length and Pro domain-cleaved KPC-1 proteins in size. The R136A, R143A mutant form with both self-cleavage sites mutated lacked the ΔPro band.
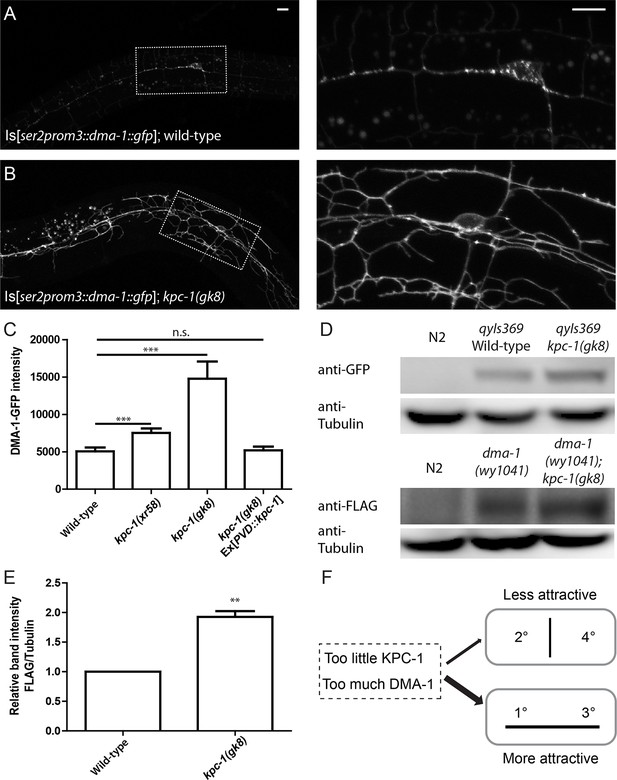
The level of the DMA-1 receptor was increased in kpc-1 mutants.
(A–B) Fluorescent images of DMA-1::GFP in PVD neurons in (A) wild-type and (B) kpc-1(gk8) mutant animals. DMA-1 showed diffuse staining in the entire dendritic arbor but was more enriched in vesicles in the cell body and 1° dendrites in wild-type PVD. Diffuse membrane localization of DMA-1::GFP was significantly increased and vesicle-like puncta were reduced in the kpc-1(gk8) mutant. The images on the right are zoomed-in views of the regions indicated by the dashed boxes. Scale bar: 10 μm. (C) Quantification of fluorescent intensity of diffuse DMA-1::GFP on the 2° dendrites. *** is p<0.001 by Student’s T-test. N=50 for each genotype. (D) Upper panels: Western blot against GFP in wild-type worms without transgene, wild-type worms expressing DMA-1-GFP and kpc-1(gk8) mutant worms expressing DMA-1-GFP. Lower panels: Western blot against FLAG in wild-type animals, dma-1(1041) mutants with 2xFLAG inserted into the dma-1 cytosolic domain of the endogenous genomic locus using CRISPR/Cas9, and dma-1(1041); kpc-1(gk8) double mutants. (E) Quantification of relative band intensity normalized to α-tubulin. ** is p<0.01 by Student’s T-test. N=4 (F) Schematic figure of the proposed model in which loss of KPC-1 caused increased membrane DMA-1, leading to defects in escaping from the high levels of ligands around the 1° and 3° dendrites.

KPC-1 caused specific down-regulation of DMA-1 receptor.
(A–B) Fluorescent images of HPO-30::GFP in (A) wild-type and (B) kpc-1(gk8) mutant PVD neurons. The level of HPO-30 on the membrane was not changed in kpc-1 mutants. Scale bar: 10 μm. (C) Quantification of HPO-30-GFP intensity on the branches. n.s. is p>0.05 by Student’s T-test. N=20 for each genotype. (D) Quantification of DMA-1-YFP fluorescent intensity of an endogenously tagged dma-1(wy1000) allele and its double mutants with kpc-1(gk8). *** is p<0.001 by Student’s T-test. N=20 for each genotype. (E) Quantification of dma-1 mRNA level relative to unc-104, snb-1, cdc-42 and tba-1 measured by qPCR. n.s. is p>0.05 by Student’s T-test. N=3 (F). Full Western blot against GFP in wild-type worms without transgenes, wild-type worms expressing DMA-1-GFP and kpc-1(gk8) mutants expressing DMA-1-GFP. No additional or absent bands were detected in the kpc-1 mutant.
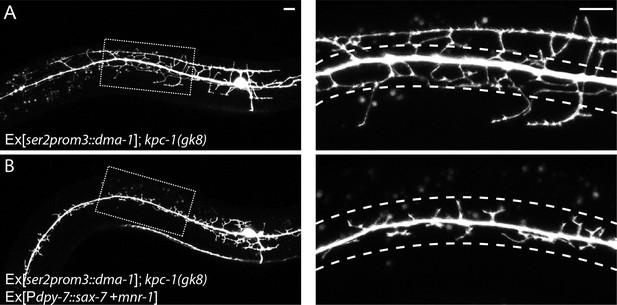
(A) Overexpressing DMA-1 caused more dendrites to escape from the trap zone.
(B) Overexpressing SAX-7 and MNR-1 in the hypodermal cells trapped the dendrites back around the 1° dendrites. The images on the right are zoomed-in views of the region indicated by the dashed boxes on the left. Dashed lines indicate the 'trap zone' with high level SAX-7. Scale bar: 10 μm.
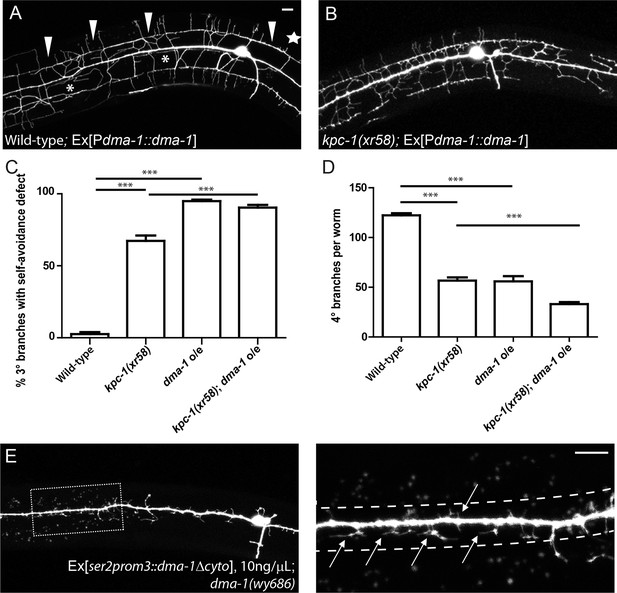
Overexpression of DMA-1 generated kpc-1 mutant-like phenotypes.
(A) Overexpression of DMA-1 in PVD neurons caused similar defects to those of the kpc-1(xr58) mutants shown in Figure 3B. Arrowheads: 3° branches that overlapped with their neighbors in mutants. Star: Defective menorah with no 4° branches. Asterisks: Trapped 2° branches. (B) Overexpressing DMA-1 in kpc-1(xr58) enhanced the 3° self-avoidance and 4° outgrowth phenotypes. (C) Quantification of the percentage of 3° branches that made contact with their neighbors. (D) Quantification of the total number of 4° branches per animal. *** is p<0.001 by Student’s T-test. N=50 for each genotype. (E) Overexpressing truncated DMA-1 without its cytosolic domain produced dramatic trapping phenotype. Arrows: Trapped dendrites. Dotted lines indicated the trap zone. Scale bar: 10 μm.

Regulation of DMA-1 by KPC-1 did not require the cytosolic domain of DMA-1.
(A) Fluorescent image showing PVD morphology of the wy908 cytosolic deleted mutant allele of dma-1. 2° and 3° dendrites were mostly intact but the number of 4° branches was reduced. (B) Dendrites of dma-1(wy908); kpc-1(gk8) double mutants were still trapped. (C) Schematic of the DMA-1 protein showing the cytosolic domain deleted in the wy908 allele. (D) Cytosolic domain-truncated DMA-1 expressed in dma-1(wy686) null mutants as a transgene at low concentration gave rise to the same phenotype as wy908. When expressed at a higher concentration, this construct caused robust trapping phenotype shown in Figure 5E. Scale bar: 10μm.
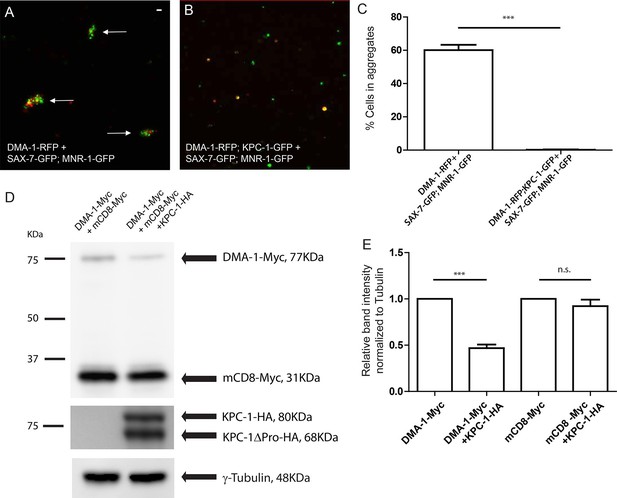
KPC-1 interrupted the interaction between DMA-1 and SAX-7/MNR-1 by down-regulating membrane DMA-1.
(A) Drosophila S2 cells co-expressing SAX-7-GFP and MNR-1-GFP formed aggregates with cells expressing DMA-1-RFP alone. (B) Cell aggregation failed when KPC-1-GFP is co-transfected with DMA-1-RFP. (C) Quantification of percentages of fluorescent cells in aggregates after 3 hr. *** is p<0.001 by Student’s T-test. The experiment was repeated three times for quantification. (D) Immunoblot showing that the amount of DMA-1 was significantly reduced when co-transfected with KPC-1 while that of another co-transfected type I transmembrane protein, mCD8, was not affected. (E) Quantification of band intensity on the Western blots. *** is p<0.001 and n.s. is p>0.05 by Student’s T-test. Each experiment was repeated three times for quantification.
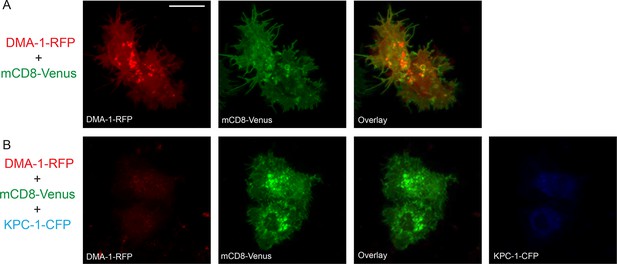
KPC-1 prevented DMA-1 from localizing to the plasma membrane.
(A) Fluorescent images showing S2R+ cells expressing DMA-1-RFP and mCD8-Venus. DMA-1 localized to the plasma membrane. (B) Co-transfection with KPC-1-CFP caused depletion of DMA-1 from the membrane while mCD8-Venus was unaffected. Scale bar: 10 μm.
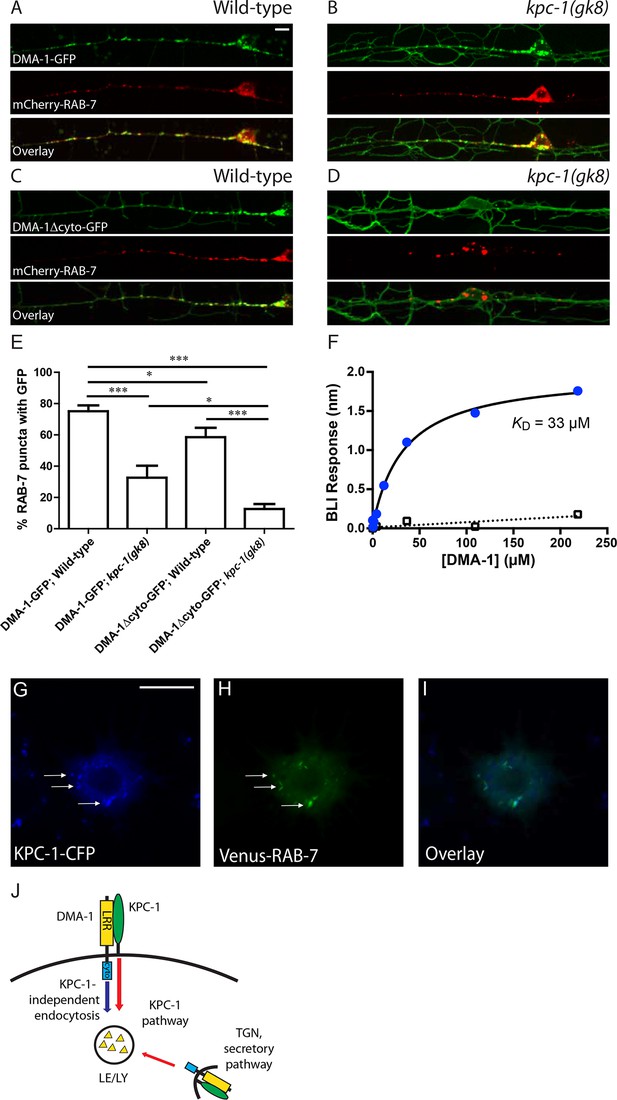
KPC-1 targeted DMA-1 to endocytic vesicles through direct interaction with its ectodomain.
(A–B) Fluorescent images of DMA-1-GFP (upper panels), mCherry-RAB-7 (middle panels) and overlay (lower panels) in PVD neurons of (A) wild-type and (B) kpc-1(gk8) mutant animals. Many DMA-1-GFP puncta co-localized with late endosomes/lysosomes labeled by mCherry-RAB-7 in wild-type PVD while the co-localization was reduced in the kpc-1(gk8) mutants. kpc-1 mutants showed enhanced DMA-1-GFP fluorescence on the membrane of dendritic branches but less in vesicles. (C–D) Fluorescent images of DMA-1Δcyto-GFP (upper panels), mCherry-RAB-7 (middle panels) and overlay (lower panels) in PVD neurons of (C) wild-type and (D) kpc-1(gk8) mutants. DMA-1 lacking its entire cytosolic domain showed brighter signal on the plasma membrane but still localized to endocytic vesicles. However, in the kpc-1(gk8) mutants, DMA-1Δcyto-GFP was almost exclusively on the plasma membrane but was absent from RAB-7-positive versicles. Scale bar: 10 μm. (E) Quantification of the percentages of mCherry-RAB-7 vesicles that showed DMA-1-GFP or DMA-1Δcyto-GFP fluorescence. *** is p<0.001, * is p<0.05 by Student’s T-test. N=20 for each genotype. (F) DMA-1 binding on KPC-1-immoilized surface using biolayer interferometry. Blue circles represent DMA-1 binding responses on KPC-1, with the black curve as the fit to a Langmuir isotherm model. Open rectangles show DMA-1 binding on a negative-control surface with the SA tip decorated with biotynlated ectodomain of human GPR56. (G-I) Fluorescent images showing the localization of (G) KPC-1-CFP, (H) Venus-RAB-7 and (I) overlay in S2R+ cells. Arrow: KPC-1-CFP puncta that co-localized with Venus-RAB-7. Scale bar: 10 μm. (J) Schematic illustration of the model. Plasma membrane DMA-1 is down-regulated via two synergistic mechanisms: KPC-1 binds to the ectodomain of DMA-1 and targets it to endosomes, while other endocytic pathways signal through the cytosolic domain of DMA-1. LRR: Leucine rich repeats domain, cyto: cytosolic domain, LE/LY: Late endosomes/lysosomes
Additional files
-
Supplementary file 1
Table S1 Mutant alleles and transgenes used in this study.
Table S2 Plasmids used in this study. Table S3 sgRNAs and repair oligos for CRISPR.
- https://doi.org/10.7554/eLife.11008.019





