The DREAM complex through its subunit Lin37 cooperates with Rb to initiate quiescence
Figures
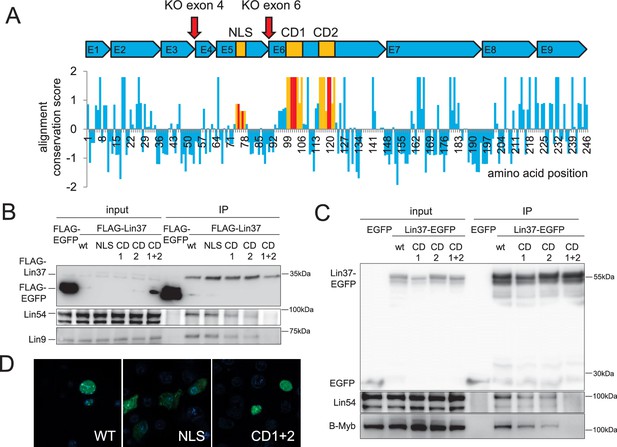
Conserved sequences in LIN37 are essential for nuclear localization and interaction with MuvB complex components.
(A) Structure and evolutionary conservation of Lin37. Conserved domains are highlighted in yellow and introduced mutations in red (NLS: K75A/R77A, CD1: D101A/R102G/S103A, CD2: C119A/R120G). Red arrows mark target sites for CRISPR/Cas9 mutagenesis. Wild-type and mutant Lin37 fused with N-terminal FLAG tags or C-terminal EGFP tags were expressed in (B) NIH3T3 cells or (C) HCT116 cells (input, 10 µg whole cell extract) and precipitated with anti-Flag antibodies or anti-EGFP antibodies (IP). The MuvB complex components Lin9, Lin54 and B-Myb were tested for interaction with Lin37 wild-type (WT) and mutants (NLS, CD1, CD2, CD1 +2). (D) Fusions of Lin37 variants with EGFP were expressed in NIH3T3 cells and tested for their subcellular localization (green). Nuclei were stained with Hoechst33342 (blue).
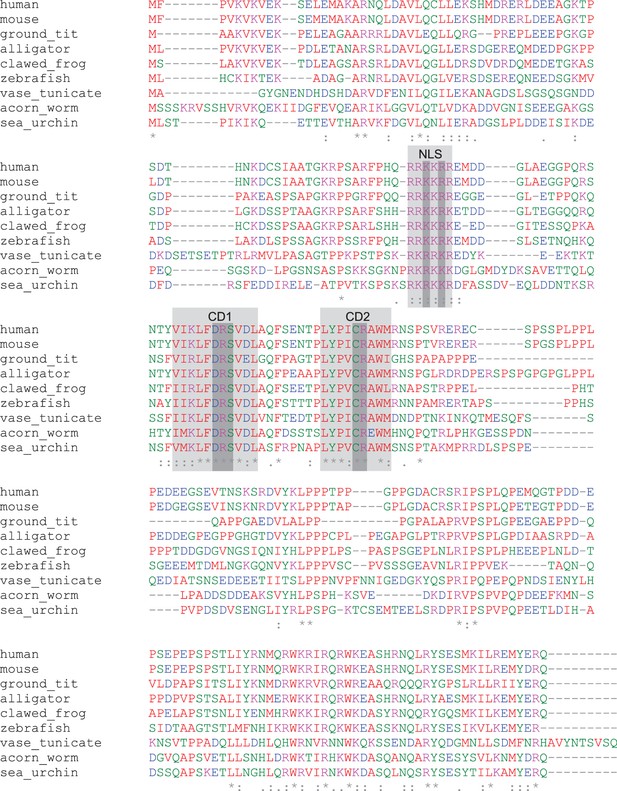
Lin37 protein is weakly conserved in deuterostomes.
An alignment of protein sequences from nine deuterostome species show that only small regions are highly conserved: one potential NLS and two domains (CD1, CD2) without predicted function. Light gray: NLS, CD1, CD2. Dark grey: amino acids mutated in mouse Lin37 to analyze functions of conserved domains.
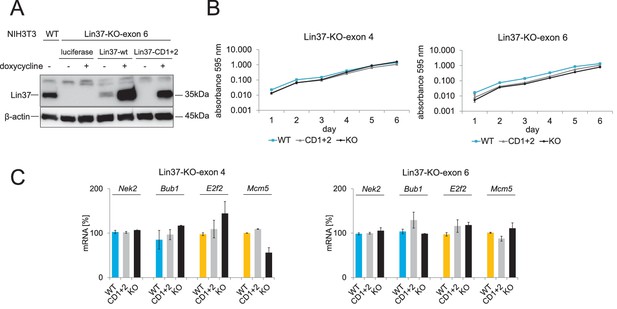
Cell viability and mRNA expression of early and late cell cycle genes in proliferating cells do not depend on Lin37.
Episomal constructs for inducible expression of Lin37 (WT), the MuvB-binding mutant (CD1 +2) and luciferase (KO) were stably introduced in Lin37 knockout clones mutated in exon 4 or exon 6. (A) Lin37-KO-exon 6 cells were tested for doxycycline-induced expression by Western blot. (B) The viability of two independent Lin37 knockout cell lines, generated by mutation of either exon 4 or exon 6, was tested by MTT assays for 6 days after stable transfection with episomal rescue constructs. Mean values ± SD of three biological replicates are shown. (C) Expression of G2/M (blue) and G1/S (yellow) cell cycle genes in proliferating cells was analyzed by qPCR. Mean values ± SD of one representative experiment for all cell lines with two technical replicates are given. All data points for each gene were normalized to the mRNA expression of the first replicate in wild-type cells which was set to 100%.
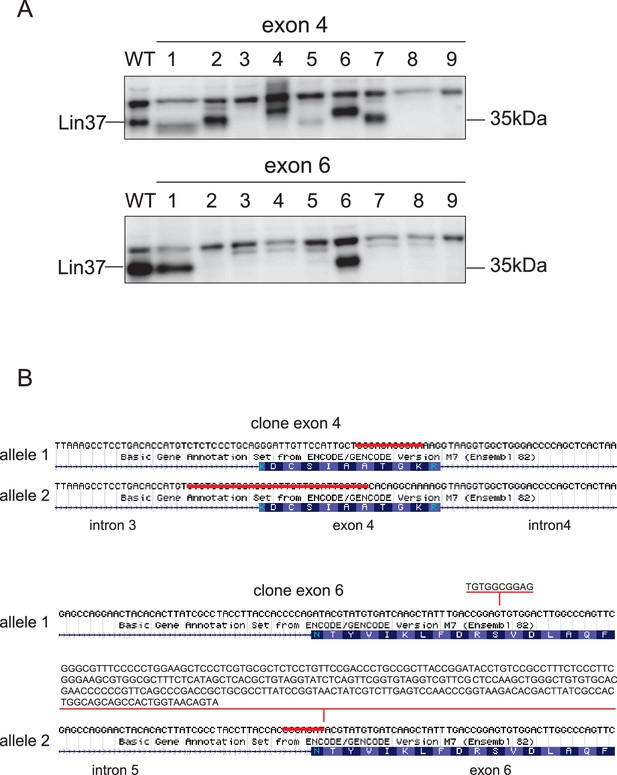
Lin37 knockout clones can be established by CRISPR/Cas9 nickase with a high efficiency.
RIPA lysates of Lin37 wild-type NIH3T3 and single cell clones treated with plasmids expressing sgRNA targeting the 5’ end of Lin37 exon 4 or 6 were probed with a Lin37 antibody (A). Nine representative clones are shown for each approach. (B) Genotyping of two clones showing deletions (clone exon 4) or insertion and deletions (clone exon6) in the Lin37 gene introduced by CRISPR/Cas9 nickase.
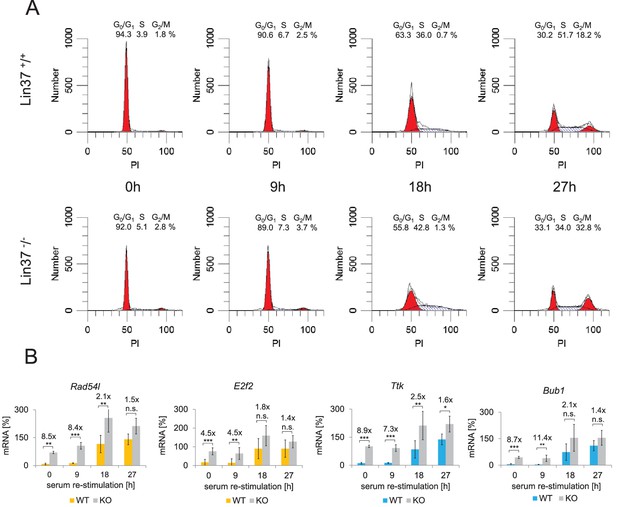
Knockout of Lin37 results in a de-repression of cell cycle genes in G0/G1.
The NIH3T3 parental cell line as well as independent wild-type (WT, n = 4) and Lin37 knockout (KO, n = 6) single cell clones were arrested in G0 by serum deprivation and re-stimulated for 9, 18, and 27 hr. (A) Cell cycle distribution was measured by PI staining and flow cytometry. One representative example of wild-type (Lin37+/+) and knockout (Lin37−/−) cells is shown. (B) mRNA of G1/S (yellow) and G2/M (blue) cell cycle genes was measured by qPCR in all cell lines with two technical replicates each and normalized to U6 expression. All data points for each gene were further normalized to the maximum mRNA expression of the parental cell line which was set to 100%. Mean values ± SD are given, and significances were calculated by the Students T-Test (*p≤0.05, **p≤0.01, ***p≤0.001).
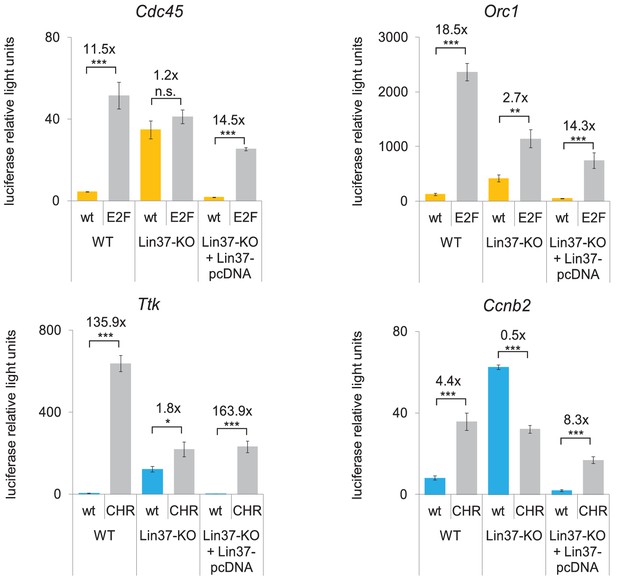
Knockout of Lin37 leads to a de-repression of cell cycle gene promoters in quiescence.
Wild-type (WT) or Lin37 knockout cells (Lin37-KO; clone 632–2) were transfected with luciferase reporter constructs of G1/S (yellow) and G2/M (blue) cell cycle gene promoters. Wild-type (wt) and DREAM-binding site deficient (E2F, CHR) promoters were tested for their activity. Lin37 knockout was rescued by re-expression of Lin37 (Lin37-pcDNA). Cells were arrested by serum deprivation. Promoter activities were normalized to Renilla luciferase activity. Mean values ± SD of three biological replicates are given, and significances were calculated by the Students T-Test (*p≤0.05, **p≤0.01, ***p≤0.001).
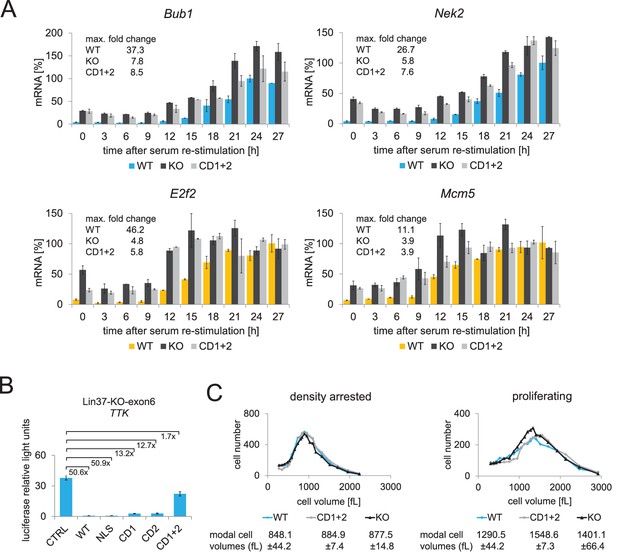
Expression of wild-type Lin37, but not of a MuvB-binding deficient mutant, rescues the knockout phenotype.
(A) Lin37 knockout cells stably transfected with episomal vectors expressing wild-type Lin37 (WT), a MuvB-binding deficient mutant (CD1+2) or luciferase (KO) were arrested by serum starvation in G0 and stimulated to re-enter the cell cycle. mRNA expression of G2/M (blue) and G1/S (yellow) cell cycle genes was measured by qPCR at given time points after serum re-stimulation. All data points for each gene were normalized to the maximum mRNA expression of the Lin37 wild-type rescue cell line which was set to 100%. Mean values ± SD of one representative experiment with two technical replicates for each time point are shown. (B) Activity of the Ttk G2/M cell cycle gene promoter in serum-starved Lin37 knockout cells (clone 632–2) was determined by luciferase reporter assays while expressing wild-type Lin37 (WT), a mutant with non-functional nuclear localization signal (NLS), or mutants with altered MuvB binding domains (CD1, CD2, CD1+2). Mean values ± SD of three biological replicates are given and compared with Ttk promoter activity in Lin37 deficient cells (CTRL). (C) The volumes of density-arrested and proliferating Lin37-deficient cells (clone 632–2) expressing wild-type Lin37 (WT), a MuvB-binding deficient mutant (CD1+2) or luciferase (KO) was determined by Coulter counter analyses. Approximately 30,000 cells were measured in two biological replicates with three technical replicates each. One representative experiment is shown and the modal cell volumes ± SD are given.
-
Figure 5—source data 1
Cell cycle analysis of synchronized Lin37−/− NIH3T3 expressing wild-type Lin37 (WT), a non-MuvB-binding Lin37 mutant (CD1+2) or luciferase (KO).
Cells were arrested in G0 by serum starvation and re-entered the cell cycle after stimulation with 20% FCS. To determine cell cycle distribution of cell populations at specific time points after re-stimulation, DNA was stained with PI and fluorescence was measured by flow cytometry. (A) Percentages of cells in G0/G1, S, and G2/M at specific time points after re-stimulation. (B) DNA content as analyzed with ModFit LT 5.0. One representative experiment is shown.
- https://doi.org/10.7554/eLife.26876.009
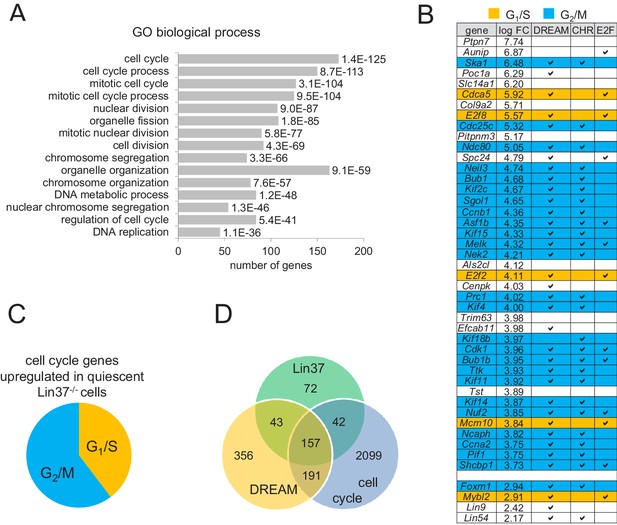
Expression of early and late cell cycle genes is up-regulated in quiescent Lin37−/− cells on a global level.
(A) Genes up-regulated in quiescent Lin37−/− cells were analyzed for enrichment of Gene Ontology (GO) terms. The top fifteen overrepresented GO terms and the number of genes are given according to the p-value. For a complete list see Supplementary file 2. (B) Top 40 genes up-regulated (log fold change) in quiescent Lin37−/− cells and de-repressed genes coding for MuvB complex components are shown. Blue: Maximum expression in G2/M. Yellow: Maximum expression in G1/S. White: No experimentally validated differential cell cycle expression. A ‘✓’ marks genes bound by DREAM components as detected by ChIP-on-chip analysis (Litovchick et al., 2007) and marks the presence of evolutionary conserved CHR or E2F binding sites close to the transcription start sites (Müller et al., 2014). For a complete list of all Lin37-regulated genes see Supplementary file 2. (C) Distribution of early and late cell cycle genes in all cell cycle genes regulated by Lin37. (D) Correlation of genes identified as regulated by Lin37, bound by DREAM and being differentially expressed during the cell cycle (Müller et al., 2014).
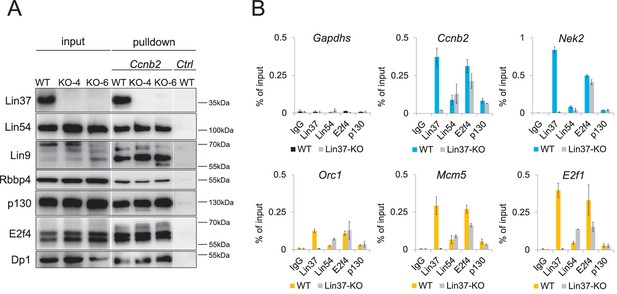
Lin37 is not essential for DREAM assembly at promoters.
(A) DREAM components were purified from nuclear extracts of two independent density-arrested Lin37 knockout cell lines created by targeting exon 4 (KO-4) or exon 6 (KO-6) and NIH3T3 cells expressing Lin37 (WT). Purification was performed with a fragment of the mouse cyclin B2 promoter (Ccnb2) containing a CDE/CHR tandem element or a fragment of the Gapdhs promoter without CHR or E2F sites (Ctrl) to determine background binding. Protein binding was analyzed by Western blotting. All samples detected with one specific antibody were run on the same gel. (B) In vivo binding of DREAM to G2/M (blue) and G1/S (yellow) cell cycle gene promoters in the Lin37+/+ NIH3T3 parental cell line (WT) and a Lin37-/- cell line (Lin37-KO; clone 632-2) was analyzed by chromatin immunoprecipitation followed by semi-quantitative PCR (ChIP-qPCR). Enrichment of DREAM components normalized to input DNA is given. Non-targeting IgG and analysis of the non-DREAM binding Gapdhs promoter served as negative controls. Mean values ± SD of one representative experiment with three technical replicates are shown.
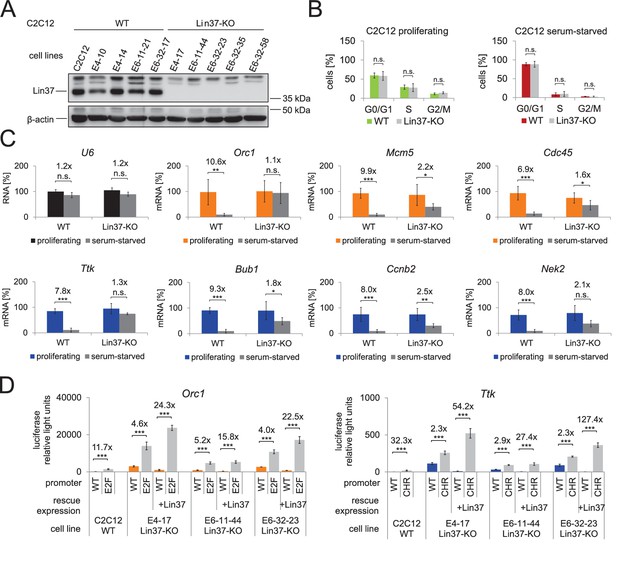
Knockout of Lin37 in C2C12 myoblast cells de-represses cell cycle genes in quiescence.
(A) Western blot analysis of Lin37 wild-type C2C12 parental cells and single cell clones treated with plasmids expressing Cas9 nickase and sgRNAs targeting the 5’ end of Lin37 exon 4 or 6. Cell extracts were tested for expression of Lin37 and β-actin. The five wild-type (WT) and Lin37 knockout (Lin37-KO) lines were utilized for further phenotypic analysis. (B) Lin37 wild-type (WT, n = 5) and Lin37 knockout (Lin37-KO, n = 5) lines were arrested in G0 by serum deprivation. Cell cycle distribution of proliferating and quiescent cells was measured by PI staining and flow cytometry. (C) mRNA expression of G1/S (orange) and G2/M (blue) cell cycle genes was measured by qPCR in all cell lines with two technical replicates each and normalized to U6 expression. All data points for each gene were normalized to the mRNA expression of the proliferating parental cell line which was set to 100%. (D) Serum-starved wild-type (WT) or Lin37 knockout cell lines (Lin37-KO) were transfected with luciferase reporter constructs of the Orc1 (G1/S) and Ttk (G2/M) cell cycle gene promoters. Wild-type (WT) and DREAM-binding site-deficient (E2F, CHR) promoters were tested for their activity. Lin37 knockout was rescued by re-expression of Lin37 (+Lin37). Promoter activities were normalized to Renilla luciferase activity. For all experiments shown in (B), (C), and (D), mean values ± SD are given, and significances were calculated by the Students T-Test (*p≤0.05, **p≤0.01, ***p≤0.001).
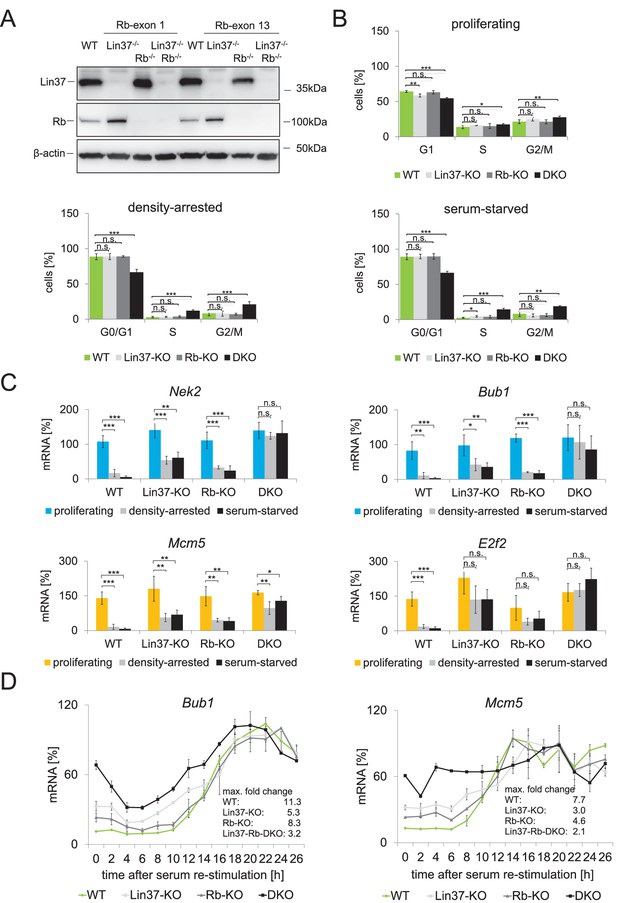
Combined ablation of Lin37 and Rb leads to loss of cell cycle control.
(A) Mutations in the Rb1 gene of NIH3T3 wild-type and Lin37−/− cells were introduced by CRISPR/Cas9 nickase by targeting exon 1 or exon 13. Loss of Rb and Lin37 protein expression was confirmed by Western blot. One representative clone for each targeting approach is shown. (B) Cell cycle distribution of proliferating, density-arrested and serum-starved wild-type (WT), Lin37−/− (Lin37-KO), Rb−/− (Rb-KO), and Lin37−/−/Rb−/− (Lin37-Rb-DKO) cell lines was analyzed by DNA-PI staining. Percentage of cells in G0/G1, S, and G2/M cell cycle phases and mean values ± SD of 4 independent cell lines each are given. Significances were calculated by the Students T-Test. (*p≤0.05, **p≤0.01, ***p≤0.001). (C) Expression of G2/M (Nek2, Bub1) and G1/S (Mcm5, E2f2) genes in the same cell lines was analyzed by qPCR. For each gene, mRNA levels were normalized to the expression of proliferating wild-type cells which were set to 100% and mean values ± SD are shown. (D) Wild-type (WT), Lin37−/− (Lin37-KO), Rb−/− (Rb-KO), and Lin37−/−/Rb−/− (DKO) cell lines were arrest by serum starvation in G0 and stimulated to re-enter the cell cycle. mRNA expression of late (Bub1) and early (Mcm5) cell cycle genes was measured by qPCR at given time points after serum re-stimulation. Mean values ± SD of one representative experiment with two technical replicates for each time point are shown. All data points for each gene were normalized to the maximum mRNA expression of the Lin37 wild-type cell line which was set to 100%. See Figure 9—source data 1 for cell cycle analysis.
-
Figure 9—source data 1
Cell cycle analysis of synchronized wild-type (WT), Lin37−/−, Rb−/−, and Lin37−/−/Rb−/− NIH3T3 cells.
To determine cell cycle distribution of serum-starved and re-stimulated cell populations, DNA was stained with PI and fluorescence was measured by flow cytometry. (A) Percentages of cells in G0/G1, S, and G2/M at specific time points after re-stimulation. (B) DNA content as analyzed with ModFit LT 5.0. One representative experiment is shown.
- https://doi.org/10.7554/eLife.26876.014
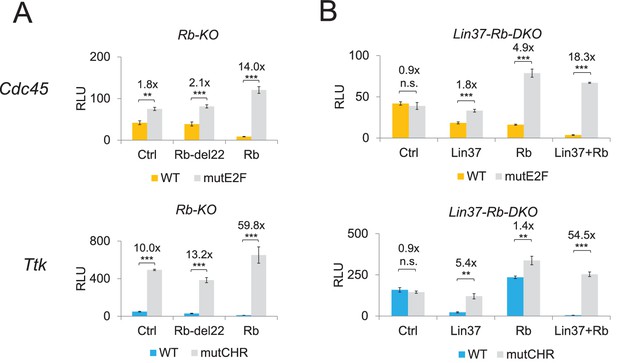
Expression of Lin37 and Rb rescues the phenotype of Rb−/− and Lin37−/−/Rb−/− cells.
Activities of wild-type (WT) and mutant (mutE2F, mutCHR) G1/S (Cdc45) and G2/M (Ttk) cell cycle gene promoters in serum-starved (A) Rb−/− (Rb-KO) and (B) Lin37−/−/Rb−/− (Lin37-Rb-DKO) cells were analyzed by luciferase reporter assays upon co-transfection of plasmids expressing Rb, a Rb mutant with non-functional pocket domain (Rb-del22), Lin37, or of an empty vector (Ctrl). Mean values of relative luciferase light units (RLU) ± SD of three biological replicates are given. Significances were calculated by the Students T-Test (*p≤0.05, **p≤0.01, ***p≤0.001).
Additional files
-
Supplementary file 1
Sequences of oligonucleotides used for cloning, mutagenesis, ChIP-qPCR, and reverse transcription qPCR.
- https://doi.org/10.7554/eLife.26876.016
-
Supplementary file 2
Transcriptome analysis of quiescent Lin37+/+ vs. Lin37−/− cells revel differentially expressed genes.
- https://doi.org/10.7554/eLife.26876.017
-
Transparent reporting form
- https://doi.org/10.7554/eLife.26876.018





