Immunogenetics: Untangling the immune basis of disease susceptibility
Why do people respond differently to the same infectious agent? A possible answer is that differences in the immune responses of individuals, caused by genetic variability between them, are responsible. Indeed, many studies have looked for associations between genes involved in immunity and disease outcome (Buniello et al., 2019), and it has been found that a gene complex called the human leukocyte antigen (HLA) system has a central role (Matzaraki et al., 2017).
The HLA genes encode the major histocompatibility complex (MHC). MHC proteins are found on the surface of cells and each one presents peptides from proteins within the cell (either proteins native to the cell or from foreign entities like bacteria) to immune cells. Thus, MHC proteins allow immune cells to recognize if a given cell is dangerous or not. For example, immune cells called CD8+ T lymphocytes recognize cells that have been infected with viruses because receptors on these T cells bind specific MHC proteins loaded with viral peptides on the surface of the infected cells. Likewise, other types of immune cells – such as natural killer cells, macrophages and dendritic cells – have receptors that bind to other parts of the MHC protein displayed by infected cells (Augusto and Petzl-Erler, 2015; Hudson and Allen, 2016).
The HLA is extremely diverse among individuals. It has been assumed that the association between some diseases and certain HLA alleles is evidence for a central role of CD8+ T cells in that disease. However, given that the HLA can also interact with receptors on other immune cells, disentangling the contributions of the different arms of the immune system remains a challenge. Now, in eLife, Becca Asquith of Imperial College and colleagues – including Bisrat Debebe as first author, researchers from various institutes in the UK and US, and the IAVI Protocol C Investigators – report the results of a new approach to disentangling these contributions (Debebe et al., 2020).
Debebe et al. reasoned that it may be possible to predict which immune cells are responsible for fighting a specific disease since receptors from particular immune cells interact with different regions of the MHC protein. For example, susceptibility to a disease may be associated with individuals carrying MHC proteins that are similar in the region that presents protein fragments to the T-cell receptor (which can be thought of as being similar in ‘T-cell receptor space’). If this is the case, then CD8+ T cells are likely involved in fighting the disease. The same reasoning can be made for associations with the other receptors, such as the killer immunoglobulin-like receptor (KIR) receptors in natural killer cells and leukocyte immunoglobulin-like receptor (LILR) in myeloid cells (Figure 1).
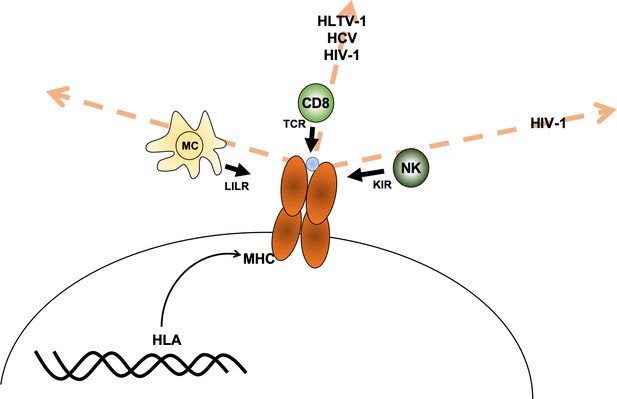
Axes of similarity in MHC proteins.
Debebe et al. showed that similarities in regions of the MHC molecule that interact with different immune cell receptors can be thought of in terms of different 'spaces' (which are defined by an axis for each type of receptor). They found that MHCs that are similar in the T-cell receptor (TCR) axis are associated with disease outcomes for HLTV-1, hepatitis C virus (HCV) and HIV-1. Disease outcomes for HIV-1 are also associated with MHCs that show similarity in the killer immunoglobulin-like receptor (KIR) axis. For the diseases tested, no MHCs were found to be similar in the region that interacts with the leukocyte immunoglobulin-like receptor (LILR) carried by myeloid cells (MC). CD8: CD8+ T cells; NK: natural killer cells; blue circle: peptide.
The challenge in this method is defining MHC similarity. Debebe et al. did so by taking an HLA known to associate with a specific disease outcome and predicting its ability to bind to different immune cell receptors. Next, they compared this predicted binding ability to that of all other HLAs, and found the HLAs with the highest binding similarity. Finally, they checked if these HLAs are also associated with disease susceptibility.
Debebe et al. applied this method to three infections (human T-cell leukemia virus type one or HTLV-1; hepatitis C virus; and HIV-1) and one autoimmune disease (Crohn’s disease). They used well-characterized cohorts where both the HLA and the outcome of the disease were known for each individual.
They studied three known HLA-outcome associations for HTLV-1 and found that other HLAs that were similar in T-cell receptor space were also associated with HTLV-1. However, other HLAs that were similar to these three HLAs in LILR space or KIR space (see Figure 1) were not associated with HTLV-1. Thus, they concluded that HTLV-1 is most likely controlled by CD8+ T cells. They also studied three HLA-outcome associations for hepatitis C, reaching the same conclusion: disease outcome is mostly controlled by CD8+ T cells. For HIV-1, Debebe et al. found that natural killer cells and CD8+ T cells both have a role, which is consistent with previous results (Bashirova et al., 2011; McBrien et al., 2018). They also studied two known HLA-outcome associations for Crohn’s disease in a large sample (2650 individuals), but other HLAs that were similar in T-cell, LILR or KIR space did not have significant effect on Crohn's disease outcome.
The approach suggested by Debebe et al. generates insights into which immune cells may be involved in fighting a disease. There are two issues that need to be explored further. First, while we know a great deal about T-cell receptors, we know less about KIR, and even less about LILR binding (Hirayasu and Arase, 2015). This knowledge (or lack thereof) is part of the definition of similarity used by Debebe et al., which can and should be updated as more data becomes available (Gwozdowicz et al., 2019). The second issue pertains to the fact that it may be possible to define similarity in other ways, leading to different results that provide further insights into how genetic variability affects the immune response.
References
-
HLA/KIR restraint of HIV: surviving the fittestAnnual Review of Immunology 29:295–317.https://doi.org/10.1146/annurev-immunol-031210-101332
-
KIR specificity and avidity of standard and unusual C1, C2, Bw4, Bw6 and A3/11 amino acid motifs at entire HLA:KIR interface between NK and target cells, the functional and evolutionary classification of HLA class I moleculesInternational Journal of Immunogenetics 46:217–231.https://doi.org/10.1111/iji.12433
-
Leukocyte Ig-like receptors - A model for MHC class I disease associationsFrontiers in Immunology 7:281.https://doi.org/10.3389/fimmu.2016.00281
-
Mechanisms of CD8+ T cell-mediated suppression of HIV/SIV replicationEuropean Journal of Immunology 48:898–914.https://doi.org/10.1002/eji.201747172
Article and author information
Author details
Publication history
- Version of Record published: May 14, 2020 (version 1)
Copyright
© 2020, Ribeiro and Graca
This article is distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use and redistribution provided that the original author and source are credited.
Metrics
-
- 2,725
- views
-
- 282
- downloads
-
- 0
- citations
Views, downloads and citations are aggregated across all versions of this paper published by eLife.
Download links
Downloads (link to download the article as PDF)
Open citations (links to open the citations from this article in various online reference manager services)
Cite this article (links to download the citations from this article in formats compatible with various reference manager tools)
Further reading
-
- Computational and Systems Biology
- Genetics and Genomics
Gene expression is known to be affected by interactions between local genetic variation and DNA accessibility, with the latter organized into three-dimensional chromatin structures. Analyses of these interactions have previously been limited, obscuring their regulatory context, and the extent to which they occur throughout the genome. Here, we undertake a genome-scale analysis of these interactions in a genetically diverse population to systematically identify global genetic–epigenetic interaction, and reveal constraints imposed by chromatin structure. We establish the extent and structure of genotype-by-epigenotype interaction using embryonic stem cells derived from Diversity Outbred mice. This mouse population segregates millions of variants from eight inbred founders, enabling precision genetic mapping with extensive genotypic and phenotypic diversity. With 176 samples profiled for genotype, gene expression, and open chromatin, we used regression modeling to infer genetic–epigenetic interactions on a genome-wide scale. Our results demonstrate that statistical interactions between genetic variants and chromatin accessibility are common throughout the genome. We found that these interactions occur within the local area of the affected gene, and that this locality corresponds to topologically associated domains (TADs). The likelihood of interaction was most strongly defined by the three-dimensional (3D) domain structure rather than linear DNA sequence. We show that stable 3D genome structure is an effective tool to guide searches for regulatory elements and, conversely, that regulatory elements in genetically diverse populations provide a means to infer 3D genome structure. We confirmed this finding with CTCF ChIP-seq that revealed strain-specific binding in the inbred founder mice. In stem cells, open chromatin participating in the most significant regression models demonstrated an enrichment for developmental genes and the TAD-forming CTCF-binding complex, providing an opportunity for statistical inference of shifting TAD boundaries operating during early development. These findings provide evidence that genetic and epigenetic factors operate within the context of 3D chromatin structure.
-
- Computational and Systems Biology
- Developmental Biology
Organisms utilize gene regulatory networks (GRN) to make fate decisions, but the regulatory mechanisms of transcription factors (TF) in GRNs are exceedingly intricate. A longstanding question in this field is how these tangled interactions synergistically contribute to decision-making procedures. To comprehensively understand the role of regulatory logic in cell fate decisions, we constructed a logic-incorporated GRN model and examined its behavior under two distinct driving forces (noise-driven and signal-driven). Under the noise-driven mode, we distilled the relationship among fate bias, regulatory logic, and noise profile. Under the signal-driven mode, we bridged regulatory logic and progression-accuracy trade-off, and uncovered distinctive trajectories of reprogramming influenced by logic motifs. In differentiation, we characterized a special logic-dependent priming stage by the solution landscape. Finally, we applied our findings to decipher three biological instances: hematopoiesis, embryogenesis, and trans-differentiation. Orthogonal to the classical analysis of expression profile, we harnessed noise patterns to construct the GRN corresponding to fate transition. Our work presents a generalizable framework for top-down fate-decision studies and a practical approach to the taxonomy of cell fate decisions.






