Rapid and sensitive detection of SARS-CoV-2 infection using quantitative peptide enrichment LC-MS analysis
Figures
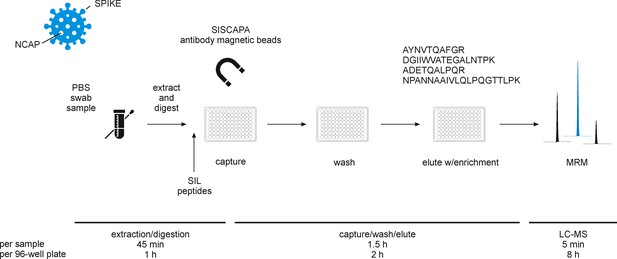
Experimental workflow for immuno-affinity peptide (stable isotope labeled [SIL] standards and capture by anti-peptide antibodies [SISCAPA]) enrichment liquid chromatography-mass spectrometry (LC-MS) of nucleocapsid protein (NCAP) severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) peptides.
Swab sample extracts were subjected to tryptic digestion, SIL standards added to the tryptic digest solution, and magnetic beads coupled with specific anti-peptide antibodies incubated to allow binding of the peptides. Unbound peptides are removed and the target peptides eluted and measured using multiple reaction monitoring (MRM) analysis with LC-MS.
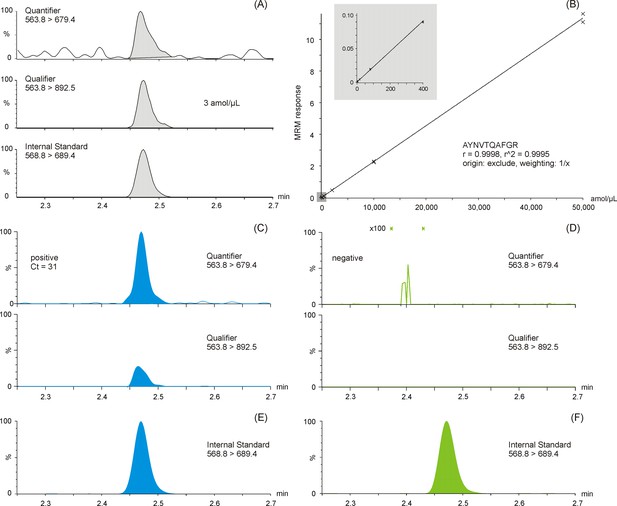
Multiple reaction monitoring (MRM) chromatograms of antibody enriched nucleocapsid protein (NCAP) AYNVTQAFGR peptide.
Quantifier, qualifier, and stable isotope labeled (SIL) internal standard peptide chromatograms spiked at the lower limit of quantification (3 amol/µl) (A). Calibration curve of the AYNVTQAFGR peptide based on enriched recombinant NCAP digest, spiked with a constant amount of SIL peptide (B). Two representative intensity-scaled MRM chromatograms of positive (mean cycle threshold [Ct] 31) (C) and negative (blank) (D) severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) swab samples, respectively, normalized to the most abundant shared MRM transition. Intensity-scaled SIL internal standard peptide MRM chromatograms of positive (E) and negative (F) SARS-CoV-2 swab samples.
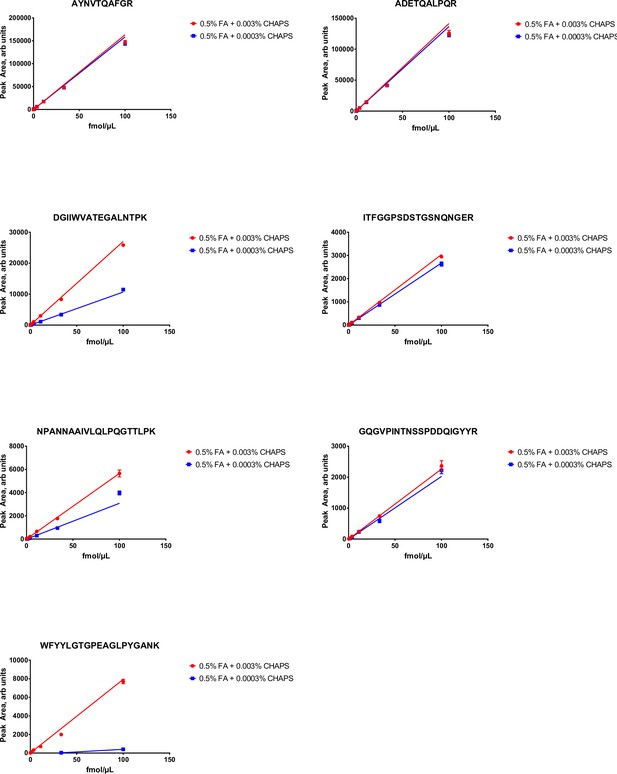
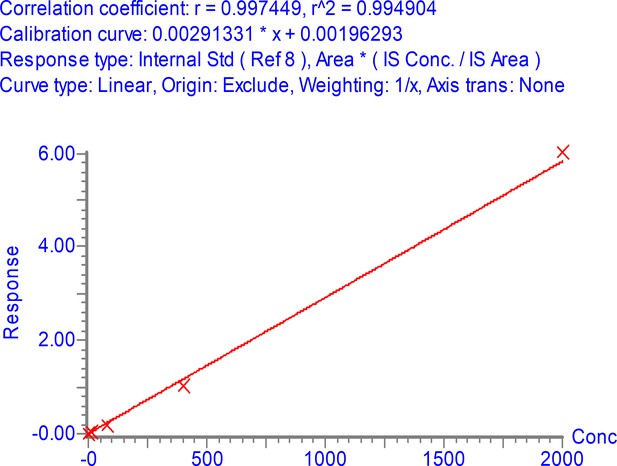
Peak area (multiple reaction monitoring [MRM] sensitivity) of stable isotope labeled (SIL) (13C615N2 C-terminal K or 13C615N4 C-terminal R labeled) nucleocapsid protein (NCAP) peptides as function of peptide and detergent (CHAPS) concentration.
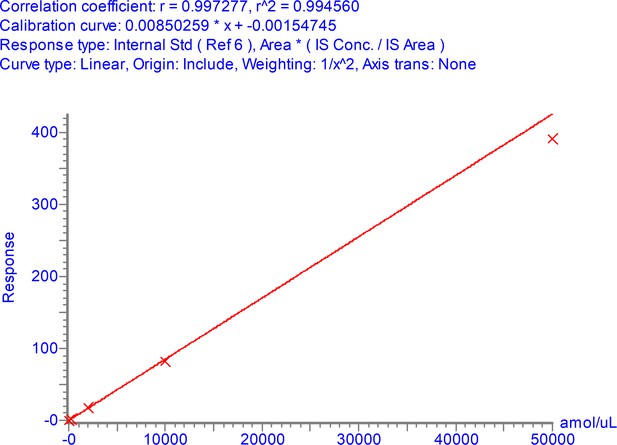
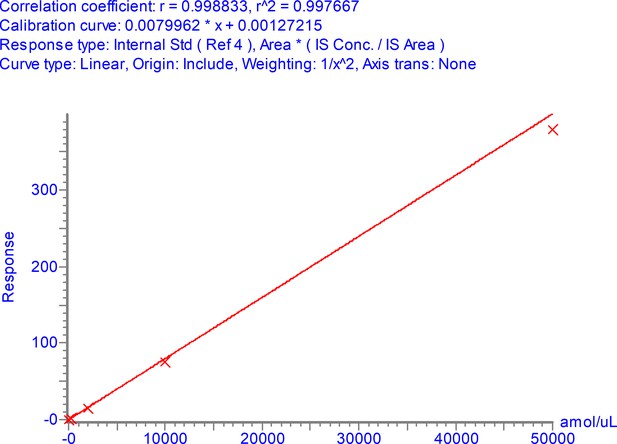
Calibration curve for NPANNAAIVLQLPQGTTLPK over the range 3–50,000 amol/µl.
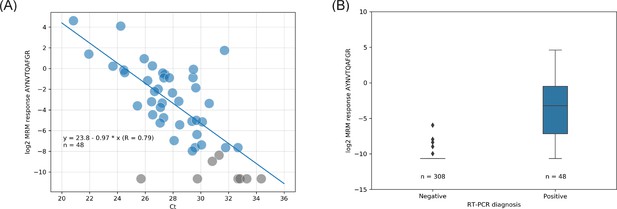
Liquid chromatography-mass spectrometry (LC-MS) (log2 quantifier response) vs. real-time polymerase chain reaction (RT-PCR) (cycle threshold [Ct]) read-out correlation with linear regression (A) and quartiles distribution of the LC-MS results (B).
Color labeling is based on RT-PCR diagnoses; blue = positive severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2); gray = not detected (no light signals) or inconclusively quantified (single transition) by LC-MS.
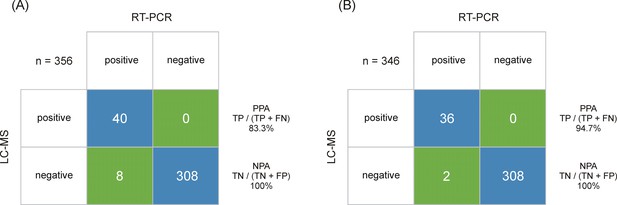
Output class (liquid chromatography-mass spectrometry [LC-MS]) vs. target class (real-time polymerase chain reaction [RT-PCR]) contingency matrix, used to calculate the positive percent agreement (PPA) and negative percent agreement (NPA) of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) immuno-affinity peptide enrichment LC-MS method (A).
The LC-multiple reaction monitoring (MRM)/MS performance is based on RT-PCR results obtained from 48 positive and 308 negative samples. (B) The LC-MRM/MS performance based on all positive samples with an RT-PCR results below cycle threshold (Ct) 30 (limit of detection [LOD] for the LC-MRM/MS) and 308 negative samples.
Tables
Intra- and inter-day method precision (n = 5) when monitoring severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) nucleocapsid protein (NCAP) peptide AYNVTQAFGR using immuno-affinity peptide enrichment liquid chromatography-mass spectrometry (LC-MS) (multiple reaction monitoring [MRM]).
| Precision (% CV) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Intra (concentration [amol/μl]) | Inter (concentration [amol/lμl]) | |||||||
| 3 | 10 | 400 | 25,000 | 3 | 10 | 400 | 25,000 | |
| Peptide-spiked PBS | 12.0 | 11.1 | 5.8 | 5.2 | – | – | – | – |
| NCAP-spiked PBS | 18.9 | 3.9 | 4.8 | 6.4 | – | – | – | – |
| Peptide-spiked VTM | 12.5 | 6.8 | 2.4 | 3.0 | 15.5 | 10.2 | 6.8 | 4.7 |
| NCAP-spiked VTM | 13.2 | 10.2 | 2.4 | 2.9 | 11.6 | 17.6 | 18.5 | 11.1 |
-
–, not tested.
Multiple reaction monitoring (MRM) transitions and mass spectrometry (MS) method details target nucleocapsid protein (NCAP) severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) peptides.
| Peptide | MRM | MRM transition type | Cone voltage (V) | Collision energy (V) | Retention time (min) | Scan window (min) |
|---|---|---|---|---|---|---|
| ADETQALPQR | 564.8 > 400.2 | Quantifier | 35 | 19 | 1.09 | 0.6–1.4 |
| 564.8 > 584.4 | Qualifier | 35 | 20 | |||
| 564.8 > 712.4 | Qualifier | 35 | 24 | |||
| 569.8 > 410.2 | SIL | 35 | 19 | |||
| AYNVTQAFGR | 563.8 > 679.4 | Quantifier | 35 | 19 | 2.49 | 2.0–3.0 |
| 563.8 > 578.3 | Qualifier | 35 | 18 | |||
| 563.8 > 892.5 | Qualifier | 35 | 19 | |||
| 568.8 > 689.4 | SIL | 35 | 19 | |||
| DGIIWVATEGALNTPK | 562.3 > 643.4 | Quantifier | 35 | 14 | 4.12 | 3.6–4.8 |
| 562.3 > 572.3 | Qualifier | 35 | 18 | |||
| 562.3 > 700.4 | Qualifier | 35 | 14 | |||
| 565.2 > 708.4 | SIL | 35 | 14 | |||
| NPANNAAIVLQLPQGTTLPK | 687.4 > 841.5 | Quantifier | 35 | 18 | 3.92 | 3.6–4.2 |
| 687.4 > 766.4 | Qualifier | 35 | 23 | |||
| 687.4 > 865.5 | Qualifier | 35 | 23 | |||
| 690.4 > 849.5 | SIL | 35 | 18 |
Additional files
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/70843/elife-70843-transrepform1-v2.pdf
-
Supplementary file 1
Integrated peak areas.
- https://cdn.elifesciences.org/articles/70843/elife-70843-supp1-v2.xlsx