Aim24 and MICOS modulate respiratory function, tafazzin-related cardiolipin modification and mitochondrial architecture
Figures
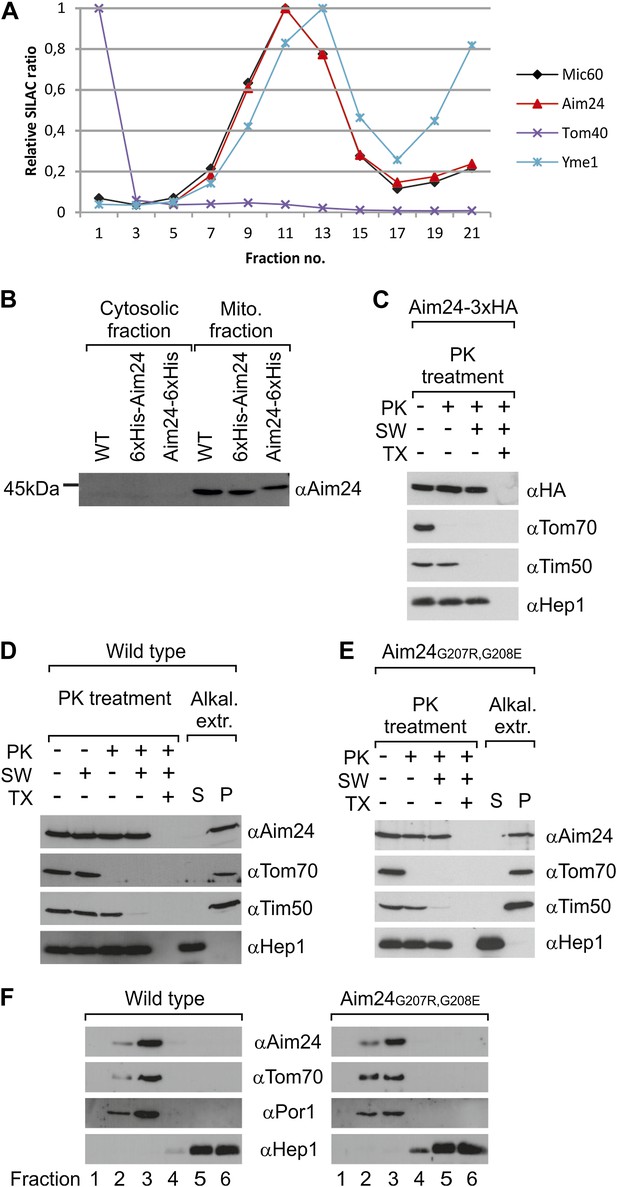
Topology of Aim24.
(A) Aim24 is a contact site protein. Isolated mitochondria from wild type (YPH499) cells were subjected to osmotic shrinking, sonication and buoyant density fractionation. Gradient fractions were analyzed by SILAC supported quantitative mass spectrometry (Harner et al., 2011). The distribution of Aim24 in the gradient is shown in red. Distribution of marker proteins of the OM (Tom40), the IM (Yme1) and of the contact site protein Mic60 are included. (B) Aim24 is synthesized as a precursor with an N-terminal targeting signal which is cleaved upon import. Wild type cells harboring pYES2 empty vector, Δaim24harboring pYES2 6xHis-Aim24 or pYES2 Aim24-6xHis were grown on SLac medium containing 0.1% glucose. Protein expression was induced by incubation for 30 min in SLac medium containing 0.1% galactose. Mitochondrial and supernatant fraction were analyzed by SDS-PAGE and immunoblotting with antibodies against Aim24.(C) The C-terminal HA-tag of Aim24-3xHA is not accessible to proteinase K (PK) added to intact mitochondria and mitoplasts. Isolated mitochondria were subjected to PK treatment, osmotic swelling (SW) and Triton X-100 (TX) as indicated and analyzed by immunoblotting. (D) Aim24 behaves like an integral membrane protein. Left: mitochondria isolated from wild type cells were treated as described in (C). Right: mitochondria were exposed to alkaline treatment at pH 12. Soluble (S) and membrane integrated (P, pellet) material was separated by centrifugation and analyzed by immunoblotting. (E) Exchange of two glycine residues in a hydrophobic stretch of Aim24 by charged residues does not alter its firm association with the membrane. Mitochondria were isolated from a Δaim24 strain harboring the pYES2 plasmid encoding Aim24 G207R, G208E and treated as described in (C). (F) Neither Aim24 wild type nor Aim24 G207R, G208E protein aggregate upon alkaline treatment of mitochondria. Mitochondria were subjected to alkaline treatment and proteins were separated by flotation gradient centrifugation. The gradients were fractionated (fractions 1–6), proteins were TCA precipitated and analyzed by immunoblotting. Fraction 1 is top and fraction 6 is bottom.
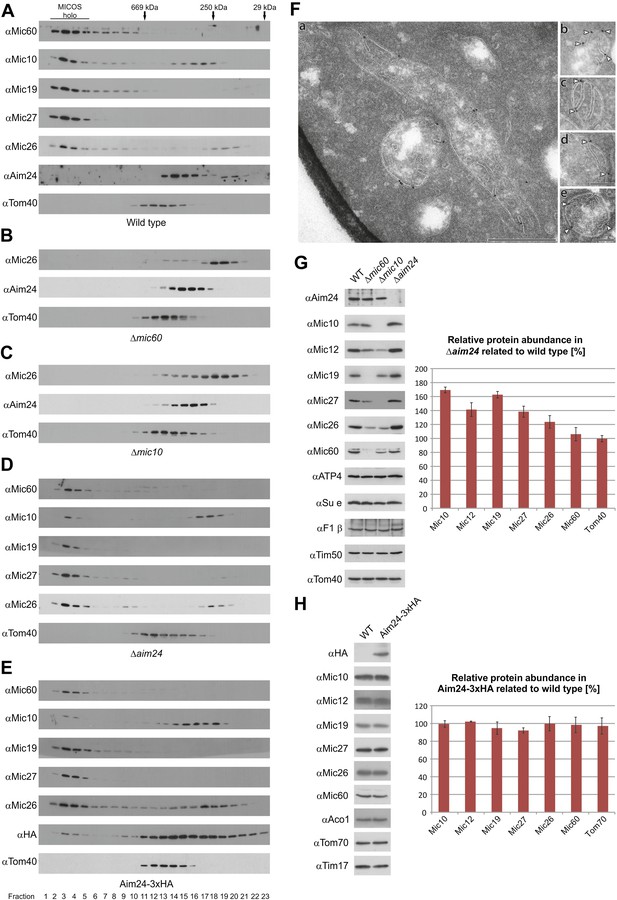
Aim24 is required for the integrity of the MICOS complex and for maintenance of steady state levels of MICOS subunits, but Aim24-3xHA is not functional.
(A–E) Integrity of the MICOS complex was analyzed by molecular sizing chromatography. Mitochondria were isolated from (A) wild type, (B) Δmic60, (C) Δmic10, (D) Δaim24 and (E) Aim24-3xHA cells. They were lysed with digitonin and subjected to gel filtration on a Superose 6 column. Proteins in elution fractions were TCA precipitated and analyzed by SDS-PAGE and immunoblotting using the indicated antibodies. The TOM complex (Tom40) was used as a control. Positions of marker proteins for calibration are indicated by arrows. Asterisks indicate a cross reaction of the Aim24 antibody used. (F, a–e) Submitochondrial localization of Mic10 by cryo-immunoelectron microscopy in a strain lacking Aim24 and harboring Mic10-3xHA. Cryo sections were labeled with anti-HA antibodies and proteinA-bound gold particles. Scale bars, 0.2 µm. Arrowheads point at gold particles at crista junctions. (G) Steady state levels of MICOS subunits in Δaim24 cells. Equal amounts of mitochondrial protein from wild type, Δmic60, Δmic10 and Δaim24 cells were analyzed by SDS-PAGE and immunoblotting using the indicated antibodies (left panel). Quantitative analysis of protein abundance of MICOS subunits and Tom40 in Δaim24 cells in relation to wild type. Error bars indicate standard deviations (right panel). (H) Steady state levels of MICOS subunits in Aim24-3xHA. Equal amounts of mitochondrial protein from wild type and Aim24-3xHA cells were analyzed by SDS-PAGE and immunoblotting using the indicated antibodies (left panel). Quantitative analysis of relative protein abundance of MICOS subunits and Tom70 in Aim24-3xHA cells. Error bars indicate standard deviations (right panel).
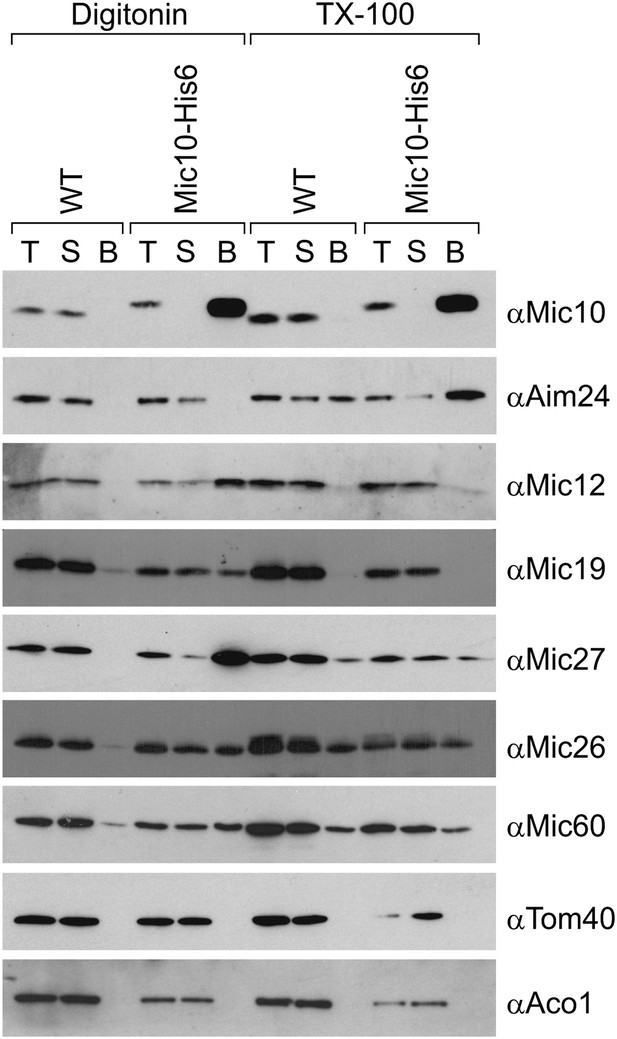
Aim24 interacts with Mic10.
Mitochondria isolated from wild type and a strain expressing Mic10-His6 were lysed with 1% digitonin or 1% TX-100 and incubated with NiNTA-beads. Bound proteins were eluted using Laemmli buffer containing 300 mM imidazole. Total (T), (5%); supernatant (S), (5%); and bound material (B), (100%) were analyzed by SDS-PAGE and immunoblotting with the indicated antibodies. Aco1, aconitase.
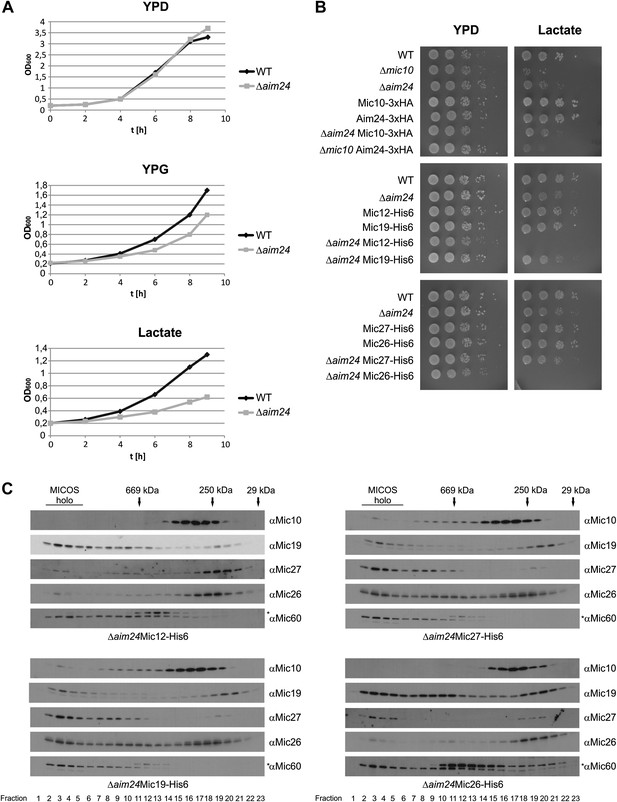
Combination of AIM24 deletion with His-tagging of Mic12 and Mic26, but not of Mic19 and Mic27, leads to respiratory deficiency and instability of the MICOS complex.
(A) Wild type and Δaim24 cells were grown in YPD, YPG or lactate liquid medium at 30°C to logarithmic phase. The cultures were diluted in the respective medium to OD600 of 0.2 and further incubated at 30°C. Aliquots were taken every 2 hr and OD600 was measured. Black, wild type; grey, Δaim24. (B) Growth of the indicated strains on agar plates containing YPD or lactate medium. Cells of the indicated strains were grown in YPD liquid medium at 30°C to logarithmic phase. Serial dilution was performed, 3 µl of each dilution was spotted on the indicated medium and incubated at 30°C. (C) Molecular sizing chromatography on Superose 6 column of the indicated strains and immunodetection of the indicated MICOS subunits. Analysis was performed as described in Figure 2A–E. Asterisks indicate a cross reaction of the Mic60 antibody used in this analysis.
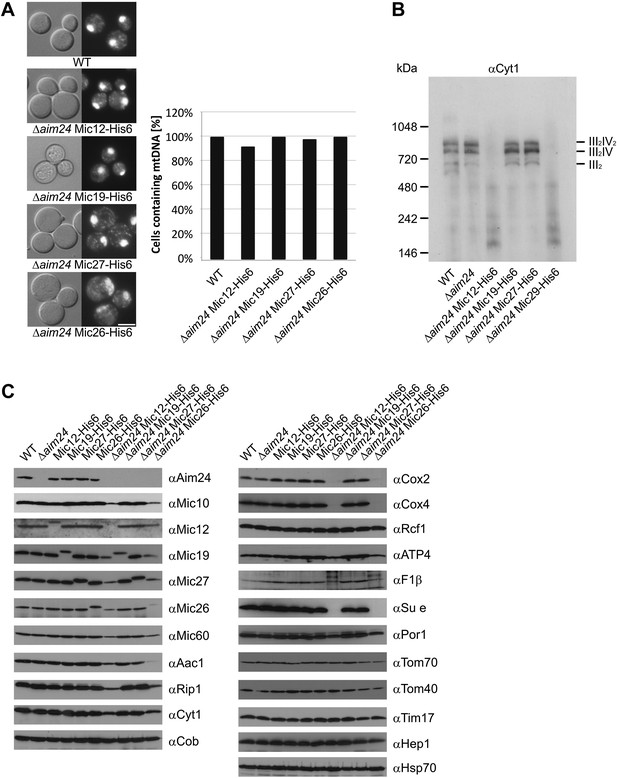
Deficiency of respiratory growth of Δaim24 mutants are not caused by lack of mitochondrial DNA, but by lack of OXPHOS supercomplexes and of a selective set of proteins involved in ATP production and transport.
(A) Fluorescence images of DAPI stained cells of the indicated strains. Scale bar, 5 µm (left panel). Quantitative analysis of levels of mitochondrial DNA by DAPI staining (right panel). (B) Analysis of respiratory supercomplexes of isolated mitochondria by Blue native gel electrophoresis and immunoblotting with antibodies against cytochrome c1 (Cyt1). The positions of supercomplexes consisting of complex III (III2) and of complexes III plus IV (III2IV and III2IV2) are indicated. (C) Steady state levels of proteins of mitochondria isolated from the indicated cells grown in the presence of lactate medium containing 0.1% glucose.
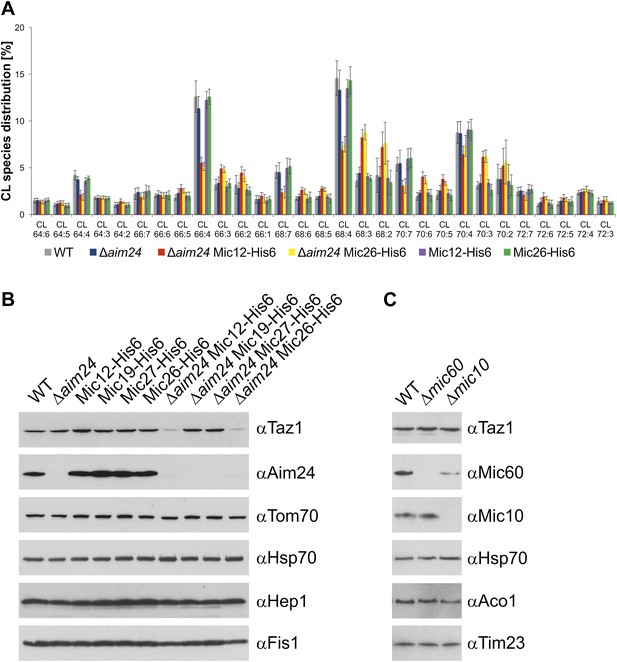
Cells lacking Aim24 and containing Mic12-His6 or Mic26-His6 are deficient in remodelling of cardiolipin and are deficient in tafazzin.
(A) Acyl chain distribution of cardiolipin. First numbers represent the sum of the carbon atoms in the four acyl chains; the numbers behind the colon indicate the total number of double bonds. Abbrevations of strains as in Figure 5. (B) Tafazzin (Taz1) is strongly down regulated in the absence of Aim24 and presence of His-tagged Mic12 or Mic26. Isolated mitochondria were analyzed by SDS-PAGE and immunodetection of the indicated proteins. (C) Presence of Taz1 is not dependent on the presence of Mic60 and Mic10. Isolated mitochondria were analyzed by SDS-PAGE and immunodetection of the indicated proteins.
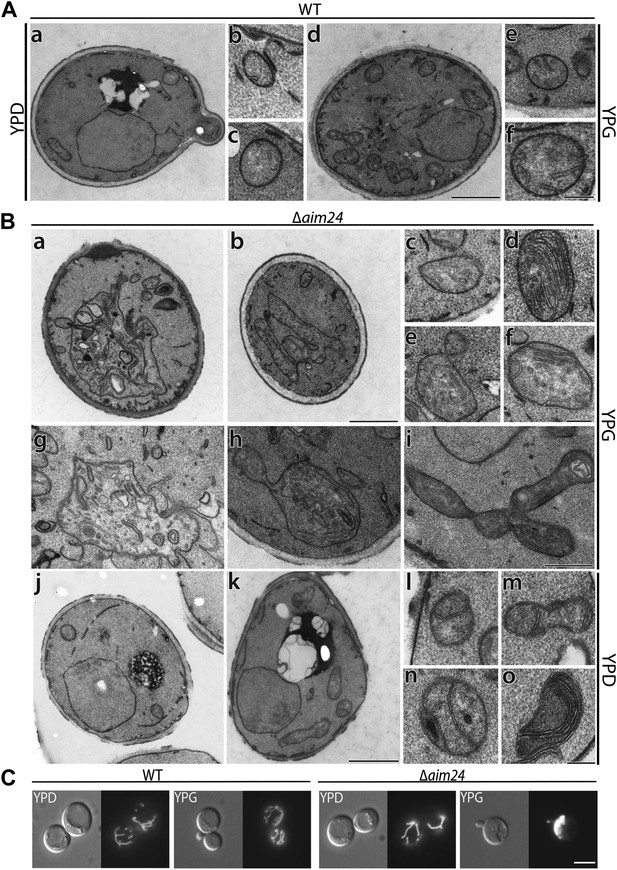
Deletion of the AIM24 gene leads to altered mitochondrial morphology and architecture.
(A and B) Electron microscopy of wild type and AIM24 deletion mutant. Cells were grown in either YPD or YPG and fixed with glutaraldehyde, contrasted with osmium tetroxide and uranylacetate. Ultrathin sections were obtained and contrasted with uranylacetate and lead citrate. (A) Wild type. a–c, cells grown on YPD; d–f cells grown on YPG; a and d, cells (scale bar, 1 µm); b, c, e, f, mitochondrial profiles (scale bar, 0.2 µm). (B) Δaim24. a–i, cells grown on YPG; j–o cells grown on YPD; a, b, j and k, cells (scale bar, 1 µm). c–i and l–o, mitochondrial profiles (scale bars, 0.2 µm). (C) Fluorescence microscopy of wild type and Δaim24 cells expressing mitoGFP. Cells were grown at 30°C on YPD or YPG medium to stationary phase (scale bar, 5 µm).
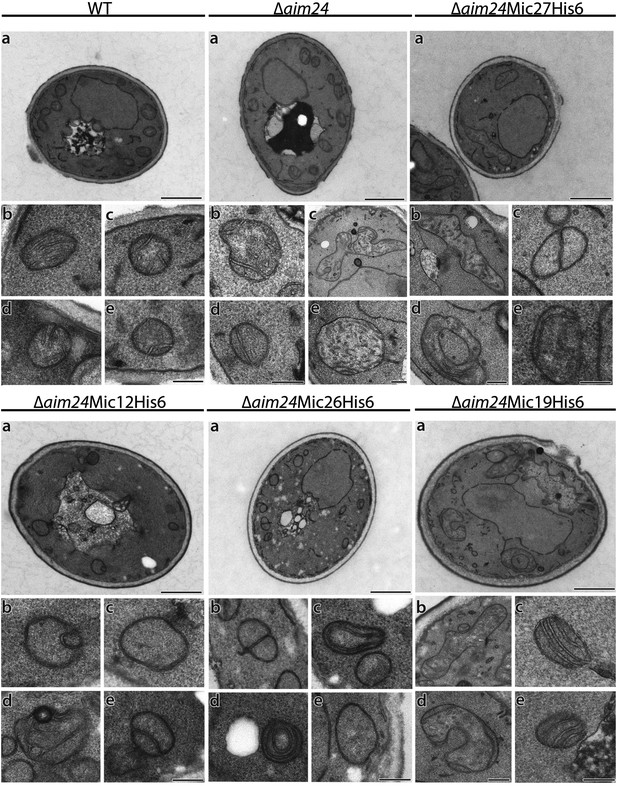
His-tagging of Mic12 and Mic26, but not of Mic19 and Mic27, in Δaim24 background leads to mitochondria grossly deficient in cristae and accumulation of septae and onion-like structures.
Cells were grown on YPG plus 0.1% glucose and electron microscopy was performed as in Figure 7A,B. a, cells (scale bars, 1 µm); b–e, mitochondrial profiles (scale bars, 0.2 µm).
Tables
Statistical analysis of the distribution of cardiolipin (CL) species in mitochondria from Δaim24 Mic12-His6 and Δaim24 Mic26-His6 cells compared to wild type
| CL species | Δaim24 Mic12-His6 | Δaim24 Mic26-His6 |
|---|---|---|
| 64:6 | ns | ns |
| 64:5 | ns | ns |
| 64:4 | * 0.015 | ** 0.0037 |
| 64:3 | ns | ns |
| 64:2 | * 0.0416 | ns |
| 66:7 | ns | ns |
| 66:6 | ns | ns |
| 66:5 | ** 0.0014 | ** 0.0030 |
| 66:4 | ** 0.0023 | ** 0.0014 |
| 66:3 | ** 0.0038 | ** 0.0047 |
| 66:2 | ns | ns |
| 68:7 | *** <0.0001 | *** 0.0002 |
| 68:6 | *** 0.0001 | *** 0.0006 |
| 68:5 | *** <0.0001 | *** <0.0001 |
| 68:4 | *** <0.0001 | *** <0.0001 |
| 68:3 | *** <0.0001 | *** <0.0001 |
| 68:2 | * 0.0130 | * 0.0156 |
| 70:7 | *** 0.0001 | *** 0.0003 |
| 70:6 | *** <0.0001 | *** <0.0001 |
| 70:5 | *** <0.0001 | *** 0.0002 |
| 70:4 | ** 0.0028 | ns |
| 70:3 | *** <0.0001 | *** <0.0001 |
| 70:2 | ns | ns |
| 72:7 | ns | * 0.0480 |
| 72:6 | ** 0.0013 | *** <0.0001 |
| 72:5 | ** 0.0055 | ** 0.0057 |
| 72:4 | ns | * 0.0359 |
| 72:3 | ns | ns |
-
Mitochondria from cells of the respective strains grown on lactate medium containing 0.1% glucose were isolated. Lipids were extracted and analyzed by mass spectrometry. Analyses contain three biological and technical replicates. The first numbers of CL species represent the sum of the carbon atoms in the four acyl chains; the numbers behind the colon indicate the total number of double bonds. Statistics were calculated by an unpaired t test using GraphPadPrism; two-tailed p-values with a 95% confidence interval are given with *p<0.05, **p<0.01, ***p<0.001.
Quantitative analysis of ultrastructural parameters of wild type cells (strain YPH499) and cells in which AIM24 was deleted
| Strains and media | WT, YPD | WT, YPG | Δaim24, YPD | Δaim24, YPG |
|---|---|---|---|---|
| Mitochondrial profiles | 115 | 302 | 126 | 170 |
| Profiles with a diameter >0,66 µm [%] | 8,7 | 10,3 | 19,0 | 34,7 |
| Profiles with a diameter >1,33 µm [%] | 1,7 | 1,0 | 4,0 | 15,9 |
| Septae [%] | 4,3 | 3,6 | 24,6 | 7,6 |
-
Cells were grown on glucose (YPD) or glycerol (YPG) containing media. 50 cells were randomly selected, the number and diameter of mitochondrial profiles determined as well as the number of septae. The values were normalized to 100 mitochondrial profiles (%).
Quantitative analysis of ultrastructural parameters of cells deficient in Aim24 and carrying one of the MICOS subunits Mic12, Mic19, Mic27 or Mic26 in His-tagged form
| Strains | WT | Δaim24 | Δaim24 Mic12-His6 | Δaim24 Mic19-His6 | Δaim24 Mic27-His6 | Δaim24 Mic26-His6 |
|---|---|---|---|---|---|---|
| Mitochondrial profiles | 246 | 182 | 133 | 214 | 202 | 184 |
| Profiles with a diameter >0,66 µm [%] | 5,7 | 13,7 | 1,5 | 10,8 | 7,9 | 2,7 |
| Profiles with a diameter >1,33 µm [%] | 0,8 | 5 | 0,0 | 3,3 | 1,5 | 1,6 |
| Septae [%] | 1,2 | 14,3 | 22,6 | 16,8 | 15,8 | 26,1 |
| Onion like [%] | 0 | 0,6 | 7,5 | 0,5 | 0,5 | 7,6 |
| Without cristae [%] | 14,2 | 9,9 | 61,7 | 7,9 | 6,4 | 53,8 |
| Crista junctions [%] | 68,7 | 65,4 | 3,8 | 49,5 | 53,5 | 2,2 |
-
Cells were grown on YPG supplemented with 0.1% glucose to allow comparison of the cells with His-tagged Mic12 and Mic26 which are unable to grow on YPG. 50 cells of each strain were randomly selected. The number of mitochondrial profiles, the diameter of mitochondrial profiles, the number of septae, onion-like structures, mitochondria without internal membranes and the number of crista junctions were determined. The values were normalized to 100 mitochondrial profiles (%).
S. cerevisiae mutant strains generated for this study
| Strain | Genotype |
|---|---|
| Δaim24 | MATα, ura3-52, lys2-801amber, ade2-101ocre, trp1-Δ63, his3-Δ200, leu2-Δ1, aim24::HIS3 |
| Aim24-His6 | MATα, ura3-52, lys2-801amber, ade2-101ocre, trp1-Δ63, his3-Δ200, leu2-Δ1, Aim24-His6-KAN |
| Aim24-3xHA | MATα, ura3-52, lys2-801amber, ade2-101ocre, trp1-Δ63, his3-Δ200, leu2-Δ1, Aim24-3xHA- HIS3 |
| Δaim24 Mic10-3xHA | MATα ura3-52, lys2-801amber, ade2-101ocre, trp1-Δ63, his3-Δ200, leu2-Δ1, aim24::HIS3, Mic10-3xHA-KAN |
| Δaim24 Mic12-His6 | MATα, ura3-52, lys2-801amber, ade2-101ocre, trp1-Δ63, his3-Δ200, leu2-Δ1, aim24::HIS3, Mic12-His6-KAN |
| Δaim24 Mic19-His6 | MATα, ura3-52, lys2-801amber, ade2-101ocre, trp1-Δ63, his3-Δ200, leu2-Δ1, aim24::HIS3, Mic19-His6-KAN |
| Δaim24 Mic27-His6 | MATα, ura3-52, lys2-801amber, ade2-101ocre, trp1-Δ63, his3-Δ200, leu2-Δ1, aim24::HIS3, Mic27-His6-KAN |
| Δaim24 Mic26-His6 | MATα, ura3-52, lys2-801amber, ade2-101ocre, trp1-Δ63, his3-Δ200, leu2-Δ1, aim24::HIS3, Mic26-His6-KAN |
-
All chromosomal manipulations were performed by homologous recombination using the indicated marker cassettes. Left column, names of the various strains; right column, genotypes of the respective strains.





