The transcription factor Pou3f1 promotes neural fate commitment via activation of neural lineage genes and inhibition of external signaling pathways
Decision letter
-
Hideyuki OkanoReviewing Editor; Keio University School of Medicine, Japan
eLife posts the editorial decision letter and author response on a selection of the published articles (subject to the approval of the authors). An edited version of the letter sent to the authors after peer review is shown, indicating the substantive concerns or comments; minor concerns are not usually shown. Reviewers have the opportunity to discuss the decision before the letter is sent (see review process). Similarly, the author response typically shows only responses to the major concerns raised by the reviewers.
Thank you for sending your work entitled “Oct6 promotes neural commitment via activation of neural lineage genes and inhibition of BMP/Wnt signals” for consideration at eLife. Your article has been favorably evaluated by a Senior editor, a Reviewing editor, and 3 reviewers, one of whom, Ali Brivanlou, has agreed to reveal his identity.
The Reviewing editor and the reviewers discussed their comments before we reached this decision, and the Reviewing editor has assembled the following comments to help you prepare a revised submission.
In this manuscript, the authors analyzed the role of Oct6 in during neuronal commitment and differentiation by using mouse ES cell differentiation system. The three reviewers' comments are mostly positive for this manuscript. However, to be acceptable, we would urge authors to make substantial revisions as summarized below.
1) Authors need to clarify what they mean by “neural commitment”. In many cases, very vague words are used in the text to describe the role of Oct6, e.g. “essential positive factor” or “critical regulator”, etc.
2) At the beginning of the paper, while authors are referring some microarray time course data, such data are not represented in Figure 1a. Furthermore, no reference was added in the text. Authors need to specify this.
3) While authors claim that Oct6 expression is detectable in undifferentiated ES cells and increases during differentiation. Then, what is the role of Oct6 in ES cells? Does Oct6 bind to the Wnt/BMP target genes and repress them?
4) Controls for overexpression and KD experiments are not clear. Authors need to clarify it. Furthermore, the effects of Oct6 KD and overexpression are relatively small upon gene expression profile and cell fate than expected. How do such relatively small changes translate to the role of Oct6 in neural induction by targeting both intrinsic transcription factors and extracellular signaling molecules? Authors need to explain about these results.
5) In the Oct6 KD experiment, there is no information about the level of KD. It is necessary to show how much the shRNAs to Oct6 is down-regulating the Oct6 gene at the transcript and protein level.
6) Do Oct-family members compensate for the depletion of Oct6?
7) It also needs to be shown that constitutive and dox-inducible Oct6 overexpression in the mouse ES cells in within the physiological range at the transcript and protein levels.
8) The Oct6 target genes identified in gain-of-function lines should be validated in the loss-of-function lines.
9) Multiple media conditions are used in various parts of the manuscript during neural differentiation. For example, data in Figure 1 is not compatible to data from other figures. Ideally, the data for all figures should be presented in serum free conditions.
10) As regards the results of Oct6 loss-of-function experiments are not consistent with those obtained in the previous report (Iwabuchi-Doi et al., 2012). Such differences should be explained in the text.
https://doi.org/10.7554/eLife.02224.019Author response
1) Authors need to clarify what they mean by “neural commitment”. In many cases, very vague words are used in the text to describe the role of Oct6, e.g. “essential positive factor” or “critical regulator”, etc.
To clarify the description, we replaced “neural commitment” with “neural fate commitment” in the revised manuscript (e.g., page 2, line 29). Neural fate commitment is an essential step in neural induction, which is defined as the process by which cells differentiate to the neural fate even in the presence of inhibitory signals (Wilson and Edlund, 2001). In addition, we also removed the words mentioned by the reviewers, such as “essential positive factor,” and “critical regulator” from the revised manuscript.
2) At the beginning of the paper, while authors are referring some microarray time course data, such data are not represented in Figure 1a. Furthermore, no reference was added in the text. Authors need to specify this.
To identify the intrinsic factors that are involved in the neural differentiation of pluripotent stem cells, we performed microarray assays in mouse ESCs undergoing neural differentiation. ESCs were aggregated as embryonic bodies in 8% knockout serum replacement medium from days 0 to 6 (Zhang et al., 2010). RNA was extracted from ESCs at days 0, 2, 4, and 6 samples, and analyzed using Agilent Whole Mouse Genome Oligo 4X44K microarrays. The microarray data have not been published yet, and we have uploaded the data to the Dryad (doi:10.5061/dryad.3vk1g). Using differential gene expression (DEG) analysis, we identified multiple genes that were up-regulated during ESC neural differentiation, including Pou3f1.
3) While authors claim that Oct6 expression is detectable in undifferentiated ES cells and increases during differentiation. Then, what is the role of Oct6 in ES cells? Does Oct6 bind to the Wnt/BMP target genes and repress them?
We thank the reviewers for raising this interesting question. Pou3f1 expression is detectable in undifferentiated ES cells, indicating that Pou3f1 might be involved in ESC pluripotency maintenance. However, Pou3f1 expression in undifferentiated ESCs is much lower compared with differentiated ESCs at days 2 and 4 (Figure 1A and Figure 1–figure supplement 1A). In addition, the expression profiles of pluripotency markers were similar in Pou3f1 knockdown ESCs and in control ESCs (Figure 1–figure supplement 1C). Therefore, the role of Pou3f1 in ESCs is not obvious.
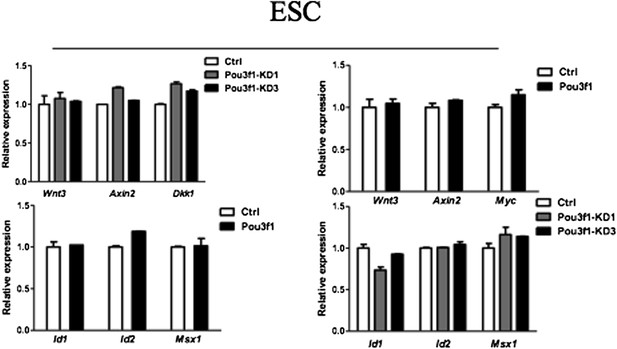
We also examined whether altering Pou3f1 expression in ESCs affects the expression of Wnt/BMP target genes. As shown in Author response image 1 below, both knockdown and overexpression of Pou3f1 have no effect on the expression of Wnt/BMP target genes in ESCs, indicating that Pou3f1 most likely does not bind to and repress the Wnt/BMP target genes in ESCs.
4) Controls for overexpression and KD experiments are not clear. Authors need to clarify it. Furthermore, the effects of Oct6 KD and overexpression are relatively small upon gene expression profile and cell fate than expected. How do such relatively small changes translate to the role of Oct6 in neural induction by targeting both intrinsic transcription factors and extracellular signaling molecules? Authors need to explain about these results.
We apologize for not making this clear in the original manuscript. For the Pou3f1-inducible overexpression experiment, we used no doxycycline addition group as control. For stable overexpression cell lines, mPou3f1 was inserted into the lentiviral expression vector pFUGW-IRES-EGFP (Naldini et al., 1996). The empty lentiviral expression vector pFUGW-GFP was used as a negative control. For the knockdown experiments, the lentiviral vector pLentiLox 3.7, which expresses a control shRNA sequence (Huang et al., 2010), was used as a negative control. We have added this information to the Materials and methods section in the revised manuscript.
For the Pou3f1 knockdown experiment, we found that the POU III family member Brn2 could compensate for Pou3f1 depletion, and we will discuss this finding in further detail in Question 6. Briefly, we found that Brn2 expression increases after Pou3f1 knockdown, whereas the simultaneous knockdown of Pou3f1 and Brn2 decreases the expression of neural markers more significantly (Figure 1–figure supplement 2D). Thus, the relatively small effects of Pou3f1 knockdown are likely due to Brn2 compensation.
As an intrinsic neural fate promoting factor, Pou3f1 is expressed earlier than the neural lineage related genes Zfp521, Pax6, and Zic1, and Pou3f1 directly binds and upregulates these genes expression (Figures 1, 4). Pou3f1-dependent direct regulation of the Sox2N2 enhancer serves as additional support for its neutralization function. These factors synergistically contribute to neuroectoderm formation (Iwafuchi-Doi et al., 2012); Pou3f1 controls their expression intrinsically and then promotes early neural fate commitment. On the other hand, the repression of extrinsic signals is essential for neural fate commitment. We demonstrate that the Pou3f1-dependent repression of BMP and Wnt signaling genes occurs at the transcriptional level (Figure 6). Pou3f1 coordinates the expression of both endogenous factors and exogenous signals at the transcriptional level, which is a fast and efficient approach to regulating cell fate determination.
5) In the Oct6 KD experiment, there is no information about the level of KD. It is necessary to show how much the shRNAs to Oct6 is down-regulating the Oct6 gene at the transcript and protein level.
We examined the effect of shRNAs to Pou3f1 on the transcript and protein levels, and found that shRNA1 (Pou3f1-KD1) and shRNA3 (Pou3f1-KD3) decreases the expression of Pou3f1 transcripts in ESCs by approximately 50% and 30%, respectively; however, shRNA2 (Pou3f1-KD2) does not affect Pou3f1 expression in ESCs (Figure 1–figure supplement 1Ba). Similar results were observed regarding the expression of Pou3f1 protein with various Pou3f1-shRNAs via Western blot (Figure 1–figure supplement 1Bb). We have added this information to the revised manuscript.
6) Do Oct-family members compensate for the depletion of Oct6?
This question is important. Oct-family members belong to POU domain factors. In previous studies, the functional redundancy between various POU factors has been reported (Andersen and Rosenfeld, 2001: Friedrich et al., 2005; Jaegle et al., 2003). Thus, we sought to address whether the compensation by other POU III family members, Brn1 and Brn2, plays a role in Pou3f1 depletion. First, we examined the expression patterns of these genes during ESC neural differentiation and observed that Brn1 and Brn2 are up-regulated in serum-free medium after day 5 (Figure 1–figure supplement 2A), which occurs much later than Pou3f1. Consistently, in mouse embryos in vivo, Pou3f1 expression is detected at E5.5, which occurs much earlier than neural induction in the mouse embryo (Figure 3A) (Zwart et al., 1996). In addition, Brn1 and Brn2 expression is only detected after E8.5 (Maxime Bouchard et al., 2005). Brn1 and Brn2 expression patterns in ESCs in vitro and mouse embryos in vivo suggest that distinct critical windows potentially exist for each POU factor to modulate neural developmental events.
Next, we assessed whether Pou3f1 depletion affects Brn1 and Brn2 expression. Indeed, Brn2 expression is obviously up-regulated in Pou3f1-KD1 and Pou3f1-KD3 cell lines compared with the control cells, whereas Brn1 expression is unchanged (Figure 1–figure supplement 2B). In addition, when both Pou3f1 and Brn2 were simultaneously knocked-down by co-transfection with lentiviral-mediated shRNAs, Brn1 expression was not affected (Figure 1–figure supplement 2C). Then, the expression of neural markers decreased more dramatically in the Pou3f1 and Brn2 double-knockdown cells compared with control or Pou3f1 knockdown cells (Figure 1–figure supplement 2D). This information was added to the revised manuscript. Together, these results suggest that Brn2 potentially compensates for Pou3f1 depletion.
7) It also needs to be shown that constitutive and dox-inducible Oct6 overexpression in the mouse ES cells in within the physiological range at the transcript and protein levels.
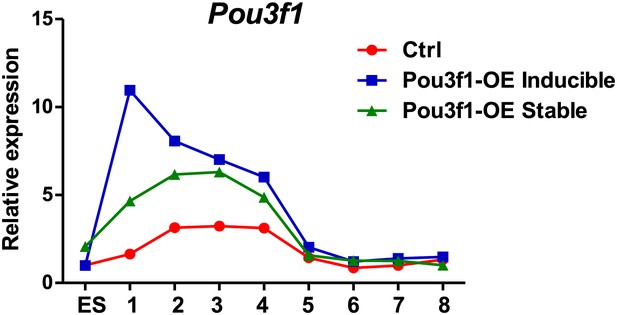
The gene functional analysis is achieved by loss-of-function or gain-of-function. Typically, a gain-of-function analysis in embryos and tissues is executed through the ectopic expression of a gene within the physiological range. However, in the cultured cells, gain-of-function is performed by overexpression, which will lead the gene expression level higher than the physiological range. To answer this question, we examined Pou3f1 expression patterns during ESC neural differentiation in control, Dox-inducible and in Pou3f1-stable overexpression cell lines. We found that the Pou3f1 expression peaked at days 2-4 of ESC neural differentiation in control cells (Author response image 2), which is consistent with the data in Figure 1A and Figure 1–figure supplement 1A. During the same period, Pou3f1 expression levels in the Dox-inducible and stable lines were approximately two-fold higher compared with control cells; except that Dox-induced Pou3f1 expression displayed a considerably higher peak at day 1 (Author response image 2). After day 5, all three cell lines displayed similar Pou3f1 expression levels.
8) The Oct6 target genes identified in gain-of-function lines should be validated in the loss-of-function lines.
As suggested by the reviewers, the Pou3f1 target genes identified in gain-of-function lines have been validated in the loss-of-function lines. The expression of neural lineage-related genes, such as Sox2, Zfp521, Zic1, and Zic2, increased in Pou3f1 overexpressing ESCs (Figure 5B), and their expression decreased after Pou3f1 depletion (Figure 5A). The expression of BMP and Wnt pathway-target genes, such as Id1/2, Msx1/2, Wnt3, Axin2, Dkk1, and Myc, was reduced in Pou3f1 overexpressing ESCs (Figures 6B and 6I), and their expression increased after Pou3f1 knockdown (Figures 6A and 6H).
9) Multiple media conditions are used in various parts of the manuscript during neural differentiation. For example, data in Figure 1 is not compatible to data from other figures. Ideally, the data for all figures should be presented in serum free conditions.
In this study, we used two culture conditions for ESC neural differentiation: serum-free medium (8% knockout replacement serum) and serum-containing medium (10% FBS). In serum-free medium, the percentage of Sox+/Oct4- neural progenitor cells (NPCs) can reach 80% of total cells by day 6 (Zhang et al., 2010); these conditions are not optimal for a gain-of-function analysis of neural fate-promoting factors but are suitable for a loss-of-function analysis. In contrast, ESC neural differentiation was inhibited in serum-containing medium, which contains BMP and other neural inhibitory signals. In addition, the percentage of NPCs is considerably reduced compared with serum-free medium after 8 days of differentiation (data not shown). The serum-containing medium is more suitable for the gain-of-function analysis of neural fate-promoting factors. Thus, we performed Pou3f1 knockdown experiments in serum-free medium (Figures 1B-D) and Pou3f1 overexpression assays in serum-containing medium (Figures 1E-G). The culture condition used depends on the experimental approaches and effects. We have added this culture condition information to the revised manuscript.
As suggested by the reviewers, we also performed the Pou3f1 overexpression experiments in serum-free medium to further confirm the function of Pou3f1 in ESC neural differentiation. The results indicate that the NPC markers, such as Sox1, Pax6, and Nestin, and the neuron markers Tuj1 and Map2 were notably upregulated in the day 4 EBs (Figure 1–figure supplement 3A). The immunostaining assays also revealed an increased number of Sox+/Oct4- NPCs, Nestin+ NPCs, and Tuj1+ neurons in the Pou3f1-stable overexpressing cells at day 4 EBs compared with control cells (Figure 1–figure supplement 3B, 3C). Cells in EBs from various days were replated in N2 medium for neuronal differentiation, and a significantly increased number of Tuj1+ neurons were observed at day 4 in the Pou3f1-overexpressing ESCs (Figure 1–figure supplement 3Cd). These results suggest that ESC neural differentiation is accelerated by Pou3f1 overexpression in serum-free culture. We have added this information to the “Results” section of Figure 1 in the revised manuscript.
10) As regards the results of Oct6 loss-of-function experiments are not consistent with those obtained in the previous report (Iwabuchi-Doi et al., 2012). Such differences should be explained in the text.
In the previous report, Iwabuchi-Doi et al. found that Pou3f1 was expressed at E7.5-E7.75 in the anterior portion of mouse embryo (Iwafuchi-Doi et al., 2012), which contributes to neuroectoderm formation (Tam and Loebel, 2007). They showed that Pou3f1 overexpression promotes the expression of neural lineage marker genes, such as Sox1 and Pax6, and concluded that Pou3f1 was involved in anterior neural plate development. In fact, our observations in Pou3f1 gain-of-function assays (Figures 1E-G and Figure 1–figure supplement 3) are consistent with their findings. The discrepancy between Iwabuchi-Doi’s study and ours occurs in the Pou3f1 loss-of-function assays. Iwabuchi-Doi et al. did not report Pou3f1 knockdown results; however, we demonstrated that Pou3f1 is required for ESC neural differentiation (Figures 1B-D). Several possibilities may contribute to the discrepancy between our study and Iwabuchi-Doi’s:
First, we differentiated mouse ESCs and EpiSCs in cell aggregates (EBs) in serum-free medium, whereas Iwabuchi-Doi et al. performed EpiSC neural differentiation using monolayers in N2B27 medium. Thus, the different differentiation protocols may account for the different observations between these two studies.
Second, the POU III family member Brn2 compensates for Pou3f1 depletion in ESC neural differentiation (Figure 1–figure supplement 2); therefore, it is difficult to capture the phenotype of ESC neural differentiation after Pou3f1 knockdown.
Third, Iwabuchi-Doi et al. have studied multiple transcriptional factors to establish the transcriptional regulatory networks during the anterior neural plate development, and Pou3f1 is only one of six factors that these authors analyzed. In our study, we exclusively focused on the function and mechanism of Pou3f1 during ESC neural differentiation, and we have to confirm Pou3f1’s function in the loss-of-function experiment very carefully. Given that the exon sequence of the Pou3f1 gene is GC-rich, it is extremely difficult to knockdown its expression effectively. We tried several shRNAs against Pou3f1, but only two of the shRNAs (Pou3f1-shRNA1 and Pou3f1-shRNA3) worked. We used these two shRNAs in our study.
We have explained this discrepancy in the revised manuscript.
https://doi.org/10.7554/eLife.02224.020