Thrombospondin-4 controls matrix assembly during development and repair of myotendinous junctions
Figures
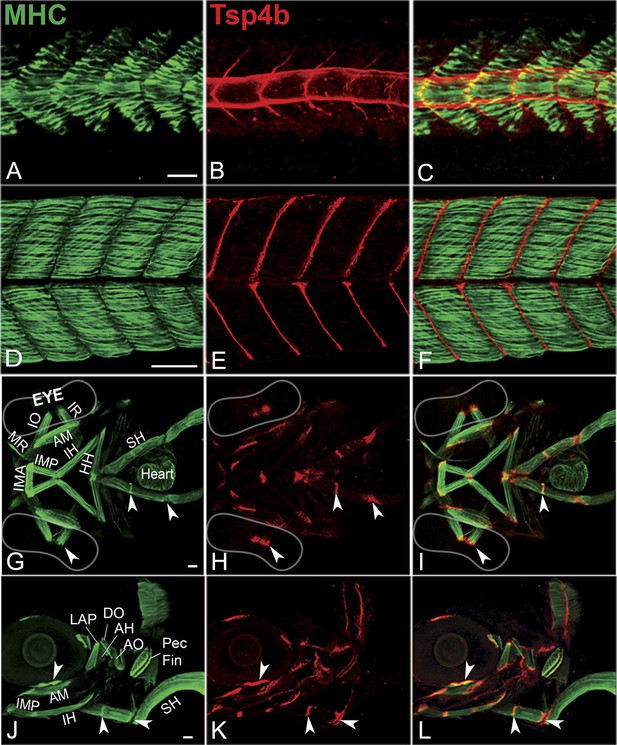
Zebrafish Tsp4b localizes to all muscle attachments.
(A–L) Whole mount immunostaining of wild type embryos using anti-MHC (A, D, G, J; green) and anti-Tsp4b (B, E, H, K; red) and merged (C, F, I, L). (A–C) 20-22 hpf and (D–F) 72 hpf (lateral view) trunk showing early Tsp4b localization around notochord and medial somite boundaries (B and C) and later at somite boundaries (E and F). (G–L) Ventral (G–I) and lateral (J–L) views of 72 hpf showing Tsp4b at cranial muscle attachments. Abbreviations: AM-Adductor Mandibularis, AH-Adductor Hyoideus, AO-Adductor Operculae, DO-Dilator Operculae, HH-HyoHyal, IH-InterHyal, IMA-InterMandibularis Anterior, IMP-InterMandibularis Posterior, IO-Inferior Oblique, IR-Inferior Rectus, LAP-LevatorArcus Palatini, MR-Medial Rectus, SH-SternoHyoideus. Scale bar = 30 microns.
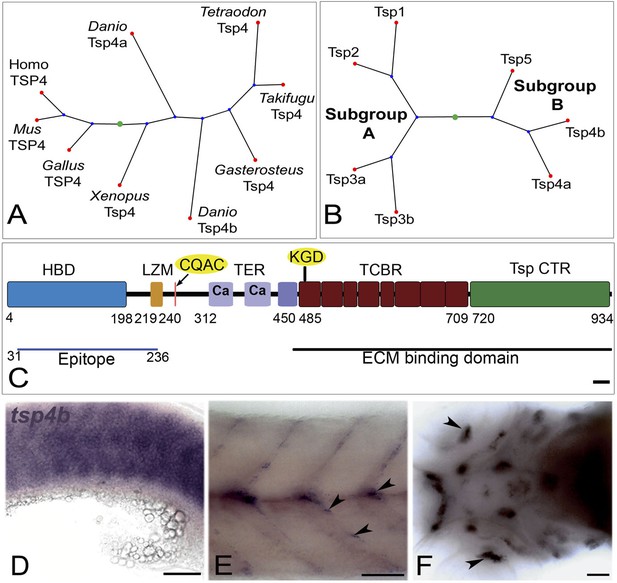
Tsp4b is expressed at muscle attachments.
(A) Phylogenetic tree showing zebrafish tsp4a and tsp4b. (B) Sequence and protein domains place Tsp4b in Thrombospondin subclass B. (C) Predicted Tsp4b functional domains: an N-terminal region (epitope aa: 31-236) containing the Heparin Binding Domain (HBD) and Leucine Zipper Motif (LZM) with Coiled-Coil Domain (CCD) with CQAS motif (261..264 aa), an integrin-binding KGD motif in the first Type 3 Calcium Binding Repeats (TCBR) domain (492-494 aa), Type II (EGF-like) Repeats (TER) and a Thrombospondin C-Terminal Region (TspCTR). Scale bar = 10 aa. (D–F) Whole mount in situ for tsp4b mRNA, anterior left, showing expression at: (D) 18 hpf (lateral view) in somites, (E) 72 hpf (lateral view) at somite boundaries. (F) 72 hpf (ventral head) at cranial muscle attachments. Scale bar = 30 microns.
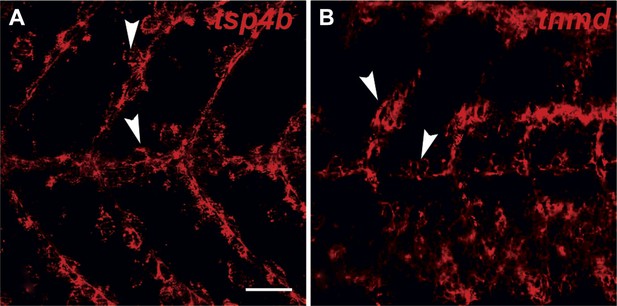
tsp4b and tnmd are expressed in tenocytes at MTJs.
(A and B) Wild type embryos at 72 hpf, labeled by fluorescent in situ hybridization and viewed laterally in whole mounts show similar patterns of expression of tsp4b (A) and tnmd (B) in groups of cells along somite boundaries (arrowheads). Scale bar = 30 microns.

Tsp4b expression is downregulated in myoblasts as they differentiate.
(A–P) Embryos triple-labeled with anti-MHC (A, green) marking differentiating myoblasts, in situ hybridization for tsp4b mRNA (B, red) and DAPI (C, blue). (A–D) Lateral views, anterior to the left, of embryos at 18 hpf show that differentiating myoblasts adjacent to the notochord lose tsp4b mRNA while expression is maintained in undifferentiated mesoderm cells, located laterally. At these stages no DAPI positive nuclei are located at somite boundaries where MHC-positive myofibers attach (arrowheads). (E–H) Optical sections of the same confocal stacks in dorsal view show absence of tsp4b expression in differentiated medial myofibers. (I–L) Lateral views of 60 hpf embryos show tsp4b mRNA localized immediately adjacent to DAPI + nuclei (dashed white ovals) of putative tenocytes along somite boundaries (arrowheads). (M–P) Optical sections in dorsal view show localized expression of tsp4b to myosepta (arrowheads). Scale bars = 35 microns.
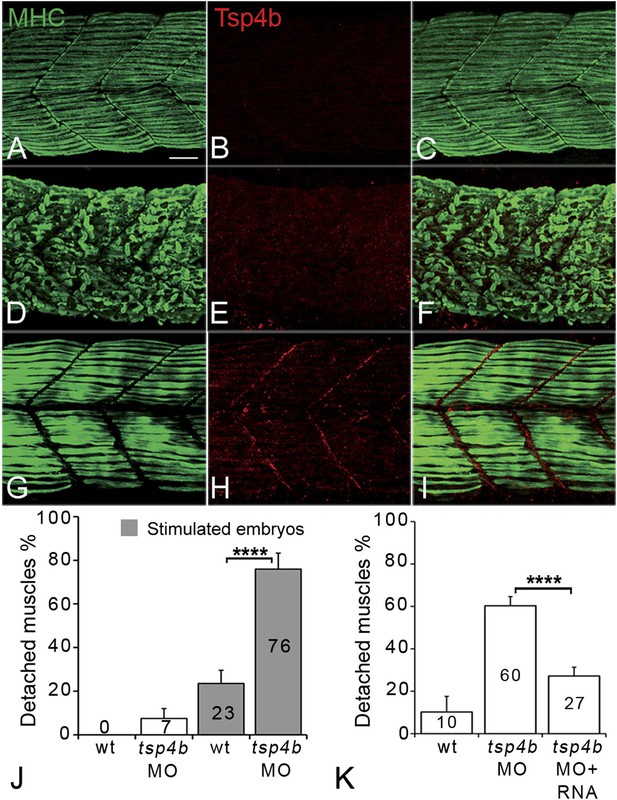
Tsp4b is required for muscle attachment.
Whole mount immunostaining of 36 hpf Tsp4b-deficient embryos using anti-MHC (A, D, G; green) and anti-Tsp4b (B, E, H; red) and merged (C, F, I). (A–C) Injection of 0.32 ng tsp4b-MO eliminates Tsp4b protein at 72 hpf but myofibers attach. (D–F) Electrical stimulation (30 V) of these larvae causes muscle detachment. (G–I) Co-injection of tsp4b RNA (80 pg/embryo) rescues muscle attachment and Tsp4b localization. (J) Histogram showing muscle detachment in 76% (N = 79) of stimulated Tsp4b-deficient embryos (Chi squared test p-value<0.001). (K) Co-injection of tsp4b mRNA rescues muscle attachment in 67% (N = 92) of stimulated Tsp4b-deficient embryos (Chi squared test p-value<0.001). (p-value representation legend: significant *<0.05, highly significant **<0.01, extremely significant ***<0.001, extremely significant ****<0.0001). Scale bar = 30 microns.
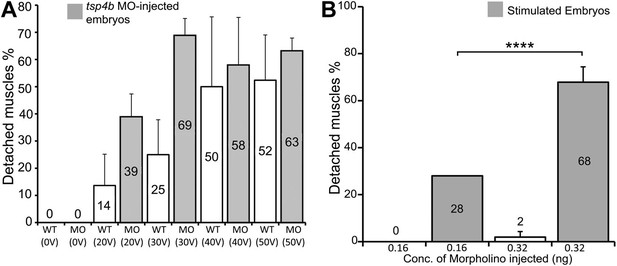
Tsp4b-deficient muscles show dose-dependent detachment upon stimulation.
(A) Histogram quantifying the percentage of embryos with at least one detached muscle fiber in wild type (white bars) and tsp4b-MO injected (gray bars) embryos subjected to mild electrical stimulation at 48 hpf to induce muscle contractions. Tsp4b-deficient embryos were more sensitive to stimulation with 30 V than 20 V, but similar amounts of detachment were observed at higher voltages, in contrast to wild-type embryos (N = 20 embryos respectively). Frequency = 4 Hz; Duration = 8 ms; Delay = 6 ms. (B) Higher tsp4b-MO doses cause more frequent detachment. Percentages of embryos with at least one detached myofiber after stimulation when injected with 0.32 ng (62.5%; N = 64) or 0.16 ng (28%; N = 64)–which reduces but does not eliminate Tsp4. (Chi squared test: p-Value<0.001).
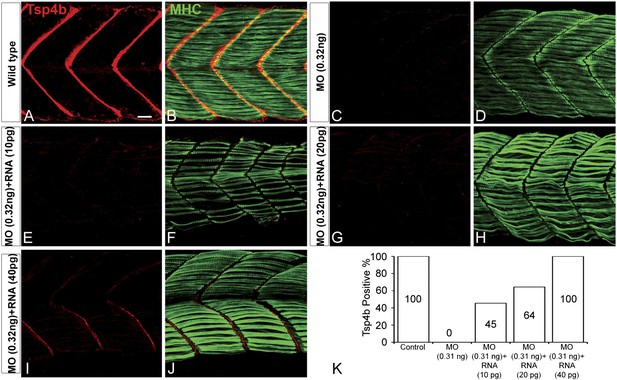
Exogenous tsp4b mRNA rescues Tsp4b localization in a dose-dependent manner.
Lateral views, anterior to the left, of somites in 48 hpf embryos stained with anti-MHC and anti-Tsp4b. (A and B) Wild types. (C and D) Tsp4b-deficient embryos injected with 0.32 ng tsp4b-MO show complete loss of Tsp4b protein. (E and F) Co-injection with 0.01 ng of full-length tsp4b mRNA restores small amounts of Tsp4b protein. (G and H) This is more pronounced with 0.02 ng of tsp4b mRNA. (I and J) 0.04 ng of tsp4b mRNA largely restores protein at somite boundaries. (K) Histogram shows that the dose-dependent increase in number of embryos with restored Tsp4b protein matches the concentration of tsp4b mRNA coinjected with tsp4b MO (N = 50 embryos). Scale bar = 30 microns.
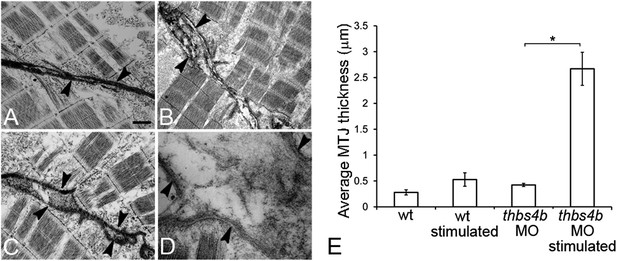
Ultrastruture of MTJs in Tsp4b deficient embryos.
(A–D) MTJ ultrastructure in wild type 72 hpf embryos before (A) and after stimulation (B), Tsp4b-deficient embryos before (C) and after stimulation (D). Arrowheads–electron-dense basement membrane (BM) at MTJs. Scale bar = 0.5 μm. (E) Histogram depicting the average spacing at MTJs, as measured in TEM images, between the BM of muscle attachments in wild type (wt) (N = 6), wild-type stimulated (N = 4), tsp4b-MO injected (N = 5), and tsp4b-MO injected and stimulated (N = 4). ‘N’ represents the number of somite boundaries analyzed in the embryos (t test one-tailed, unequal variance p-value <0.05 ANOVA posthoc Tukey test: p-value<0.001).
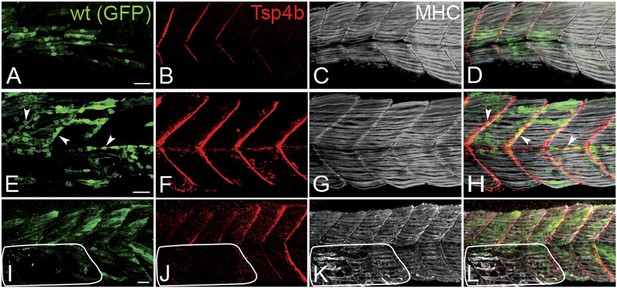
Tsp4b has a non cell-autonomous function in muscle attachments.
(A–L) 48 hpf (lateral views) of genetic mosaics generated by cell transplantation. GFP-labeled wild type muscle cells (green) (A and I) or putative tenocytes (arrow heads) (green) (E) were grafted into Tsp4b-deficient host embryos and stained with anti-Tsp4b (B, F, J; red), and anti-MHC (C, G, K; gray/white). (I–L) Transplants locally rescued muscle detachment after stimulation in regions where Tsp4b was restored (white line denotes region lacking Tsp4b). Scale bars = 30 microns.
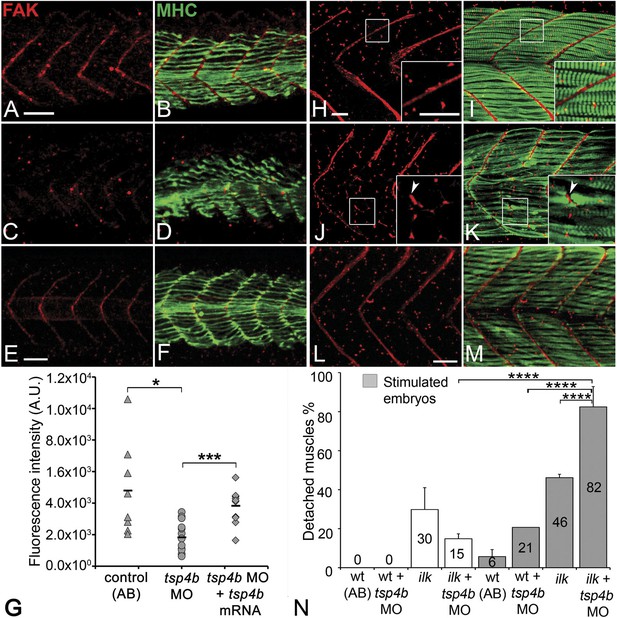
Tsp4b is required for muscle-specific integrin signaling at MTJ.
(A–F) Lateral views of 20–24 hpf embryos stained with anti-phosphorylated (Tyrosine 861) FAK (pFAK; red) and anti-MHC (green). (A and B) pFAK localizes to the ends of early myofibers at wild-type somite boundaries. (C and D) Reduced pFAK levels in Tsp4b-deficient embryos. (E and F) pFAK levels are restored in Tsp4b-deficient embryos injected with full length tsp4b mRNA. (G) Fluorescence intensity measurements (arbitrary units [A.U.]) for pFAK staining along somite boundaries confirm significant reductions in Tsp4b-deficient embryos, and partial rescue by co-injection of full length tsp4b mRNA (t test: one tailed, unequal variance; p-value: wt and tsp4b-deficient <0.05; Tsp4b-deficient and Tsp4b-deficient + tsp4b RNA p<0.001). (H–M) 36 hpf (lateral views) stained with anti-pFAK (red), and anti-MHC (green). Insets show higher magnification images of white boxed areas. (H and I) pFAK localizes to muscle sarcolemma. (J and K) In Tsp4b-deficient embryos, pFAK is reduced/discontinuous at somite boundaries. pFAK associates with ectopic muscle attachments (arrowheads). (L and M) pFAK localization is restored in Tsp4b-deficient embryos injected with full length tsp4b mRNA. (N) Embryo percentages (N = 70 embryos) with detached muscles from an intercross between two ilk+/− heterozygotes, injected with sub-threshold amounts (0.16 ng) of tsp4b-MO and stimulated (30 V) (Chi squared test; p-value: wt+ tsp4b-MO (stimulated) and ilk+tsp4b-MO (stimulated) p<0.0001, ilk (stimulated) and ilk+tsp4b-MO (stimulated) p<0.0001, ilk+ tsp4b-MO and ilk+tsp4b-MO (stimulated) p<0.0001). Scale bar = 30 microns.
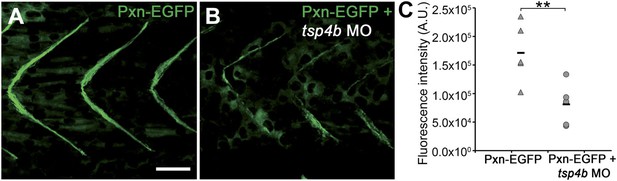
Tsp4b-deficient muscles show reduced localization of Paxillin.
(A and B) 20 hpf tg(pxn-egfp) embryo showing localization of the Pxn-EGFP fusion protein to the sarcolemma of myotubes at MTJs and reductions in embryos injected with tsp4b MO. (C) Fluorescence intensity measurements (arbitrary units [A.U.]) of Pxn-EGFP at somite boundaries confirm a significant reduction in Tsp4b deficient animals. (t test: one tailed, unequal variance; p-value<0.01) Scale bar = 30 microns.
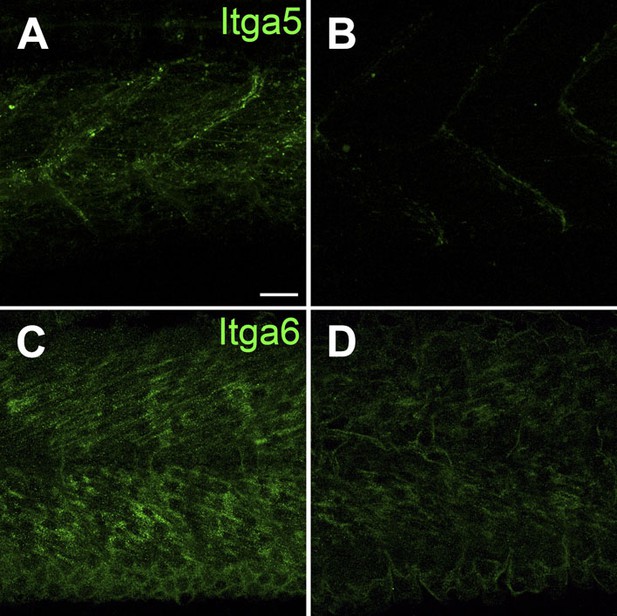
Itga5-RFP localizes to MTJs and Itga6-GFP localizes to muscle, and both require Tsp4b.
(A and B) Live imaging of embryos showing localization of tg[Itga5-RFP] in wild type (A) and Tsp4b-knockdown (B) embryos at 36 hpf. (C and D) Embryos stained with anti-GFP show localization of Itga6-GFP in wild type (C) and reduced localization in Tsp4b-knockdown samples at 36 hpf (D). Scale Bar = 20 microns.
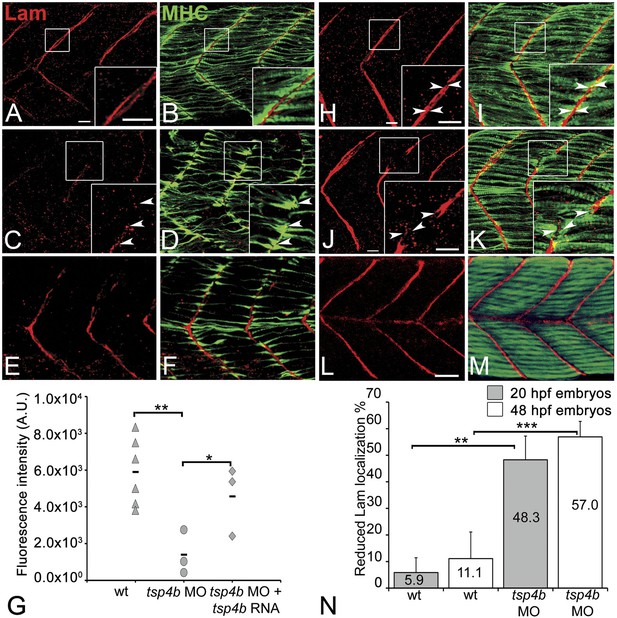
Tsp4b is required for Laminin assembly at MTJs.
(A–F) Lateral views of somites in 20 hpf embryos stained with anti-MHC (green) and anti-pan-Laminin (Lam, red) antibodies. Insets show higher magnification images of the areas marked by white boxes, where Lam localizes to developing myotendinous ECM (arrowheads). (A and B) Wild-type siblings. (C and D) tsp4b-MO injected embryos showing local loss of Lam at 20 hpf, and ectopic muscle attachments (arrowheads) at remaining Lam foci. (E and F) Lam localization was restored in Tsp4b-deficient embryos injected with full length tsp4b mRNA. (G) Fluorescence intensity measurements (arbitrary units [A.U.]) at somite boundaries of anti-Lam in 20–24 hpf wild type controls versus embryos injected with tsp4b-MO or co-injected with tsp4b-MO and tsp4b RNA. (t test: one tailed, unequal variance; p-value: wt and Tsp4b-deficient <0.01, Tsp4b-deficient and Tsp4b-deficient and tsp4b RNA <0.05) Scale bar = 30 microns. (H and I) Lateral views of somites in wild type embryos at 36 hpf stained with anti-pan-Lam (red), and anti-MHC (green). Insets show higher magnification images of white boxed areas. Lam localizes to myotendinous ECM (I, arrowheads). (J and K) In Tsp4b-deficient embryos, Lam is reduced/discontinuous at somite boundaries (K, arrowheads). (L and M) Lam localization is restored in Tsp4b-deficient embryos injected with full length tsp4b mRNA. (N) Embryo percentages (20 hpf embryos N = 30, 72 hpf embryos N = 50) with reduced/mislocalized Lam at 20 and 48 hpf in wild type and Tsp4b-deficient embryos. (Chi squared test; p-values: 20 hpf **<0.01, 48 hpf ***<0.001) Scale bar = 30 microns.

Tsp4b depletion does not alter lam transcription.
Histogram showing relative fold-changes in quantitative PCR (qRT-PCR) assays using cDNA prepared from wild-type and Tsp4b-deficient 36hpf embryos. Tsp4b-deficient embryos show no significant difference in alpha2 (lama2), beta2 (lamb2), gamma1 (lamc1) and gamma2 (lamc2), all of which are expressed in muscle. Samples are normalized against ef1alpha as a basal expression marker. rpl13a serves as an internal control.
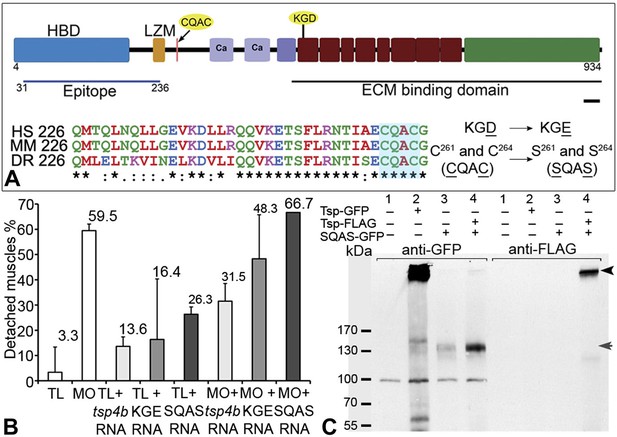
Integrin binding and pentamerization of Tsp4b are essential for its function.
(A) Schematic representation of Tsp4b domains showing the location of the conserved coiled-coil region with its CQAC motif (70% identical across species). Amino acid substitutions used to study the functions of KGD and CQAC motifs are underlined. (B) Muscle detachment frequencies in uninjected wild-type controls, wildtypes injected with tsp4b morpholino (MO), tsp4b RNA, KGE tsp4b mutant RNA, SQAS tsp4b mutant RNA, or co-injected with tsp4b MO and tsp4b RNA, KGE tsp4b mutant RNA, or SQAS tsp4b mutant RNA, after stimulation (N = 60 embryos each). (C) Western blot performed on whole embryo protein extract using anti-GFP and anti-FLAG antibodies under non-reducing conditions. Lanes: 1–wt (AB), 2–Tsp4b-deficient embryos injected with tsp4b-GFP mRNA, 3–Tsp4b-deficient embryos injected with SQAS-GFP mRNA, 4–Tsp4b-deficient embryos co-injected with tsp4b-FLAG and SQAS-GFP mRNAs. Pentameric Tsp4b-GFP (∼663 kDa) and pentameric Tsp4b-FLAG (∼535 kDa) bands (black arrow head). Monomeric SQAS-GFP (∼132 kDa) (grey arrow head). The band corresponding to a 100 kDa size marker in all lanes of the blot reacted with anti-GFP is a background signal.
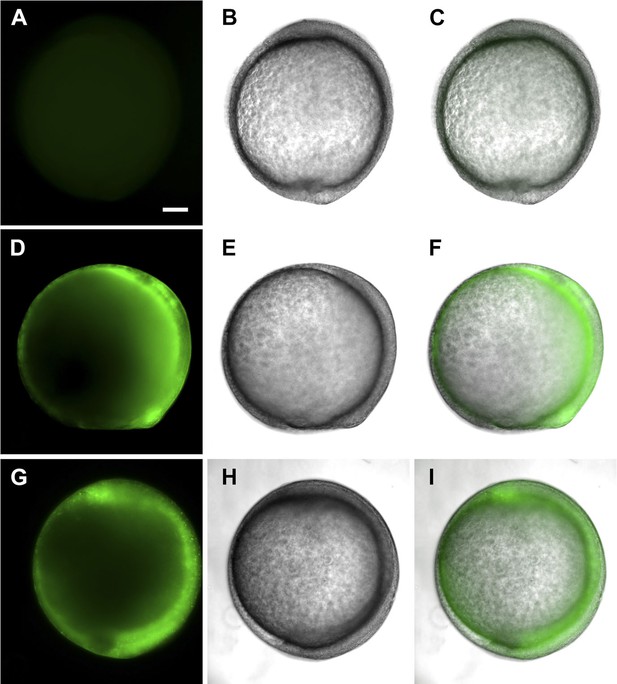
tsp4b-SQAS-gfp mRNA is expressed similar to wild type tsp4b-gfp mRNA.
(A, D, G) Live embryos at tail bud (∼10hpf) showing show GFP fluorescence. (B, E, H) embryos imaged in bright field. (C, F, I) Merged images. (A–C) Wild type embryos (D–F) Embryos injected with tsp4b-gfp transcript. (G–I) Embryos injected with SQAS-gfp mutant transcript. Scale bar = 100 microns.
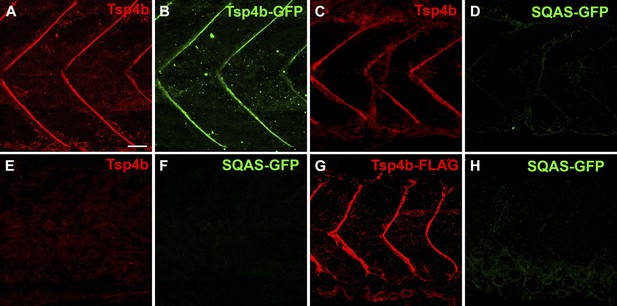
The CQAC motif is essential for Tsp4b localization and function.
(A–H) 48 hpf embryos stained with anti-GFP (green) and anti-Tsp4b (red) show localization of all Tsp4b (A) vs injected Tsp4b-GFP protein (B) in wild type embryos. (C and D) Tsp4b localizes to the MTJ (C) but injected SQAS-GFP does not (D) in wild type embryos, (E and F) Neither Tsp4b (E) nor SQAS-GFP (F) localizes to the MTJ in Tsp4b-deficient embryos. (G and H) Injected Tsp4b-FLAG (G) localizes to the MTJ while SQAS-GFP (H) does not in Tsp4b-deficient embryos (G–H). Scale bar = 20 microns.
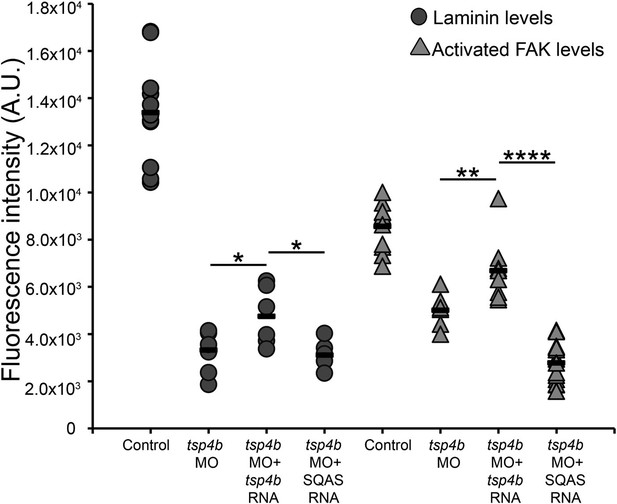
Laminin and FAK localization are dependent on pentameric Tsp4b.
Fluorescence intensity measurements (arbitrary units [A.U.]) at somite boundaries of anti-pFAK and anti-Lam immunohistochemical staining in wild type and Tsp4b-deficient embryos (48hpf), co-injected with tsp4b RNA, co-injected with SQAS tsp4b mutant RNA. (t test: one tailed, unequal variance; p-values * <0.05, ** <0.01, ****<0.0001).
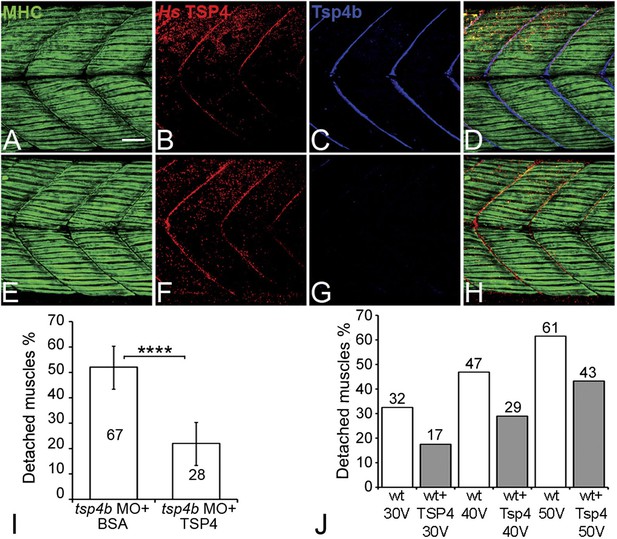
Conserved functions for TSP4 in maintenance of MTJs.
(A–H) Lateral views of trunk muscles in 72 hpf embryos stained with anti-MHC (green), anti-human TSP4 (red) and anti-Tsp4b (blue) antibodies. (A–D) Injected recombinant human TSP4 co-localizes with zebrafish Tsp4b at muscle attachments in wild type embryos. (E–H) Injected TSP4 localizes to somite boundaries in Tsp4b-deficient embryos. (I) Injected TSP4 rescues muscle attachments in Tsp4b-deficient embryos upon stimulation (N = 96 embryos) (Chi squared test, p value<0.001). (J) Histogram showing percentage of embryos with detached muscles in 60 hpf wt+BSA (white columns) and wt+TSP4 (shaded columns) embryos, stimulated at 30 V, 40 V and 50 V, respectively. N = 40 embryos for each sample. (Scale bars = 30 microns).
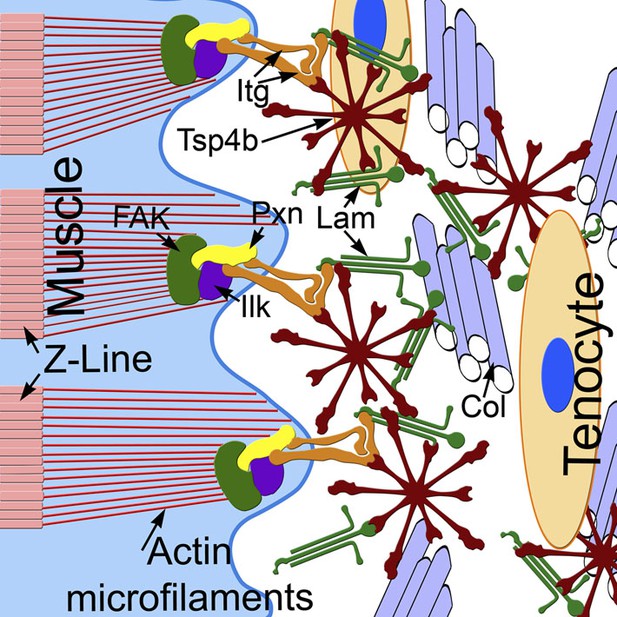
Tsp4b establishes and maintains MTJ ECM organization.
A model for ECM assembly at an MTJ. Pentameric assemblies of Tsp4b (red) associate with Lam (green), Col fibrils and other ECM components. Tsp4b and Lam bind Itgs (orange) on the muscle cell surface, activating FAK (green) and recruiting Pxn (yellow) and Ilk (purple) to promote muscle specific Itg signaling and stabilize myofiber attachment.
Additional files
-
Supplementary file 1
List of primer sequences used for qRT-PCR, cloning of mRNA and probe synthesis.
- https://doi.org/10.7554/eLife.02372.023





