LARP7 suppresses P-TEFb activity to inhibit breast cancer progression and metastasis
Figures
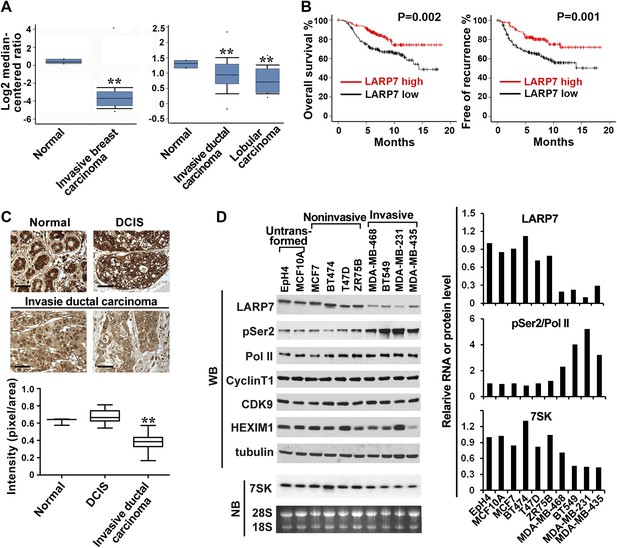
LARP7 is significantly downregulated in invasive human breast cancer tissues and cells.
(A) Box plots show decreased levels of LARP7 in invasive breast carcinoma (left), invasive ductal carcinoma, and lobular carcinoma (right) compared with normal breast tissues in two microarray data sets. **: the p values (p<0.01, compared with normal breast tissues) were determined by the Student's t test. (B) Kaplan–Meier analysis of overall survival and recurrence-free survival of breast cancer patients stratified by the expression of LARP7. The p values were calculated by the log-rank test. (C) Immunohistochemical staining of LARP7 in normal human mammary tissue (n = 6), ductal carcinoma in situ (DCIS) (n = 14), and invasive ductal carcinoma (n = 120). The intensity of LARP7 staining was quantified using ImageJ Plus and shown in the box plot below. Scale bars represent 40 μm. **: the p value (p<0.01, compared with normal breast and DCIS tissues) was determined by the Student's t test. (D) Western blotting (WB) analysis of the levels of LARP7, phospho-Ser2 (pSer2), total Pol II, CyclinT1, CDK9, and HEXIM1 in various breast cancer cell lines (upper panels) and Northern blotting (NB) analysis of 7SK snRNA levels (lower panels). Tubulin, 28S and 18S RNAs were used as loading controls. Expression of LARP7, pSer2 of Pol II, and 7SK RNA was quantified, normalized to that in EpH4 cells, and shown in the graph to the right.
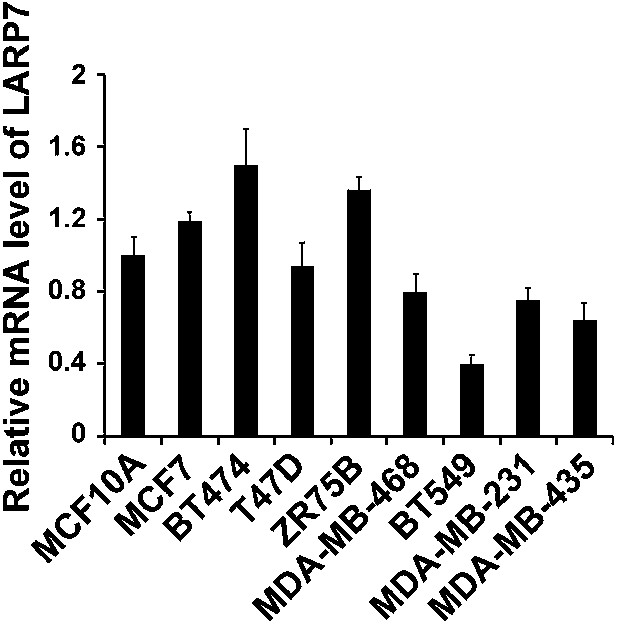
qRT-PCR analysis of LARP7 mRNA levels in untransformed MCF10A and various breast cancer cell lines.
PCR values were normalized to the levels of β-actin. Data are presented as the mean ± SD from three independent measurements.
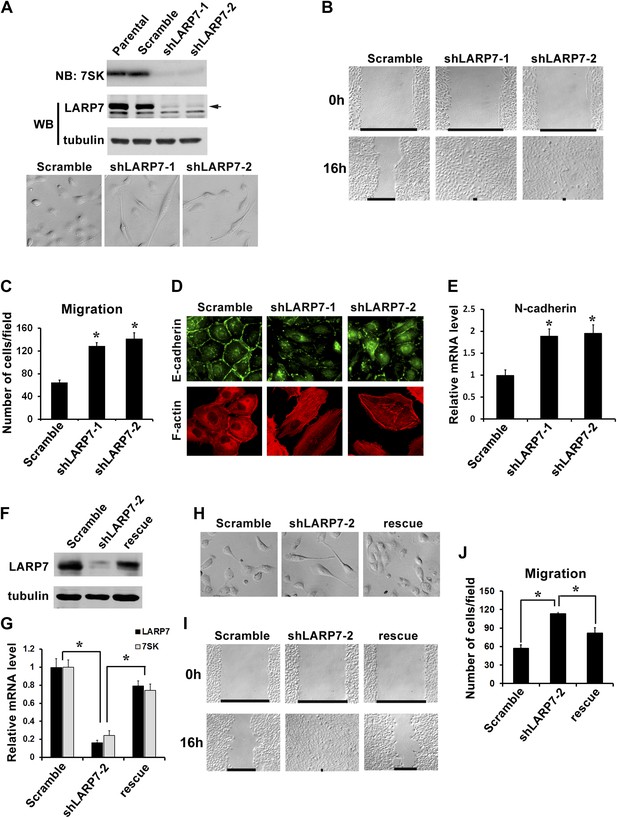
Silencing LARP7 induces EMT in MCF10A cells.
(A) Upper panel, the levels of 7SK snRNA and LARP7 protein in MCF10A parental cells and cells stably expressing scramble control, shLARP7-1, or shLARP7-2 were examined by Northern blotting (NB) and Western blotting (WB), respectively. Tubulin was used as a loading control. Lower panel, phase-contrast images of control and two shLARP7 pools. (B) Wound healing assay. Confluent cell monolayers were wounded, and wound closure was monitored at 0 hr and 16 hr. (C) Migration assay. MCF10A control or shLARP7 cells were subjected to a Transwell migration assay. The migrated cells were stained and counted. Data were collected from five fields in three independent experiments. (D) Immunofluorescence staining of E-cadherin (green) and actin stress fibers (red). (E) qRT-PCR analysis of N-cadherin expression in control and two shLARP7 pools. (F and G) The levels of LARP7 protein (F) and mRNA (G) as well as 7SK snRNA level (G) in control, shLARP7-2 cells and rescue cells (expressing an shRNA-resistant WT LARP7 cDNA) were examined by Western blotting and qRT-PCR, respectively. (H) Phase contrast pictures of control, shLARP7-2, and rescue cells. (I) The wound healing assay. (J) The Transwell migration assay. For C, E, G, and J panels: data are presented as mean ± SD. *: the p values (p<0.05) were determined by the Student's t test.
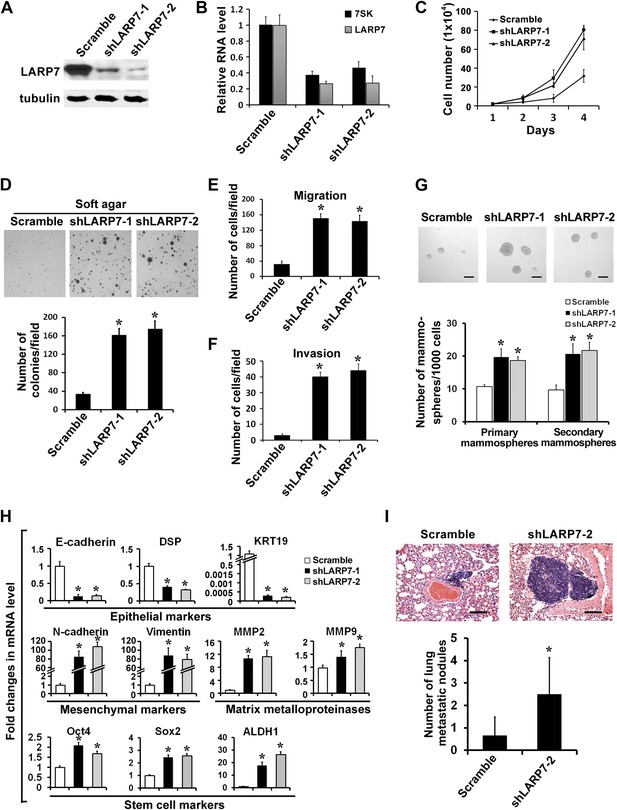
Silencing LARP7 promotes malignant progression of T47D breast cancer cells.
(A) Western blotting shows effective knockdown of LARP7 in T47D cells stably expressing shLARP7s. (B) qRT-PCR analysis of 7SK and LARP7 RNA levels in T47D cells stably expressing scramble control or shLARP7s. (C–F) Silencing LARP7 in T47D cells results in increased cell proliferation (C), anchorage-independent growth (D), cell migration (E), and invasion (F). (G) Representative images of mammospheres formed by control or shLARP7 cells. The numbers of primary and secondary mammospheres were quantified and shown in the graph below. Scale bars represent 100 μm. (H) qRT-PCR analysis of various epithelial and mesenchymal markers, MMPs, and stem cell markers in control and two shLARP7 pools. DSP, desmoplakin; KRT19, keratin 19. PCR values were normalized to that of β-actin. (I) Representative images of the H&E-stained lung sections from mice injected with control or shLARP7-2 cells. Scale bar, 100 μm. Quantification of the number of metastatic nodules is shown in the bar graph below. n = 6 in each group. Data in this figure are presented as the mean ± SD, and the p values (*p<0.05) were determined by the Student's t test.
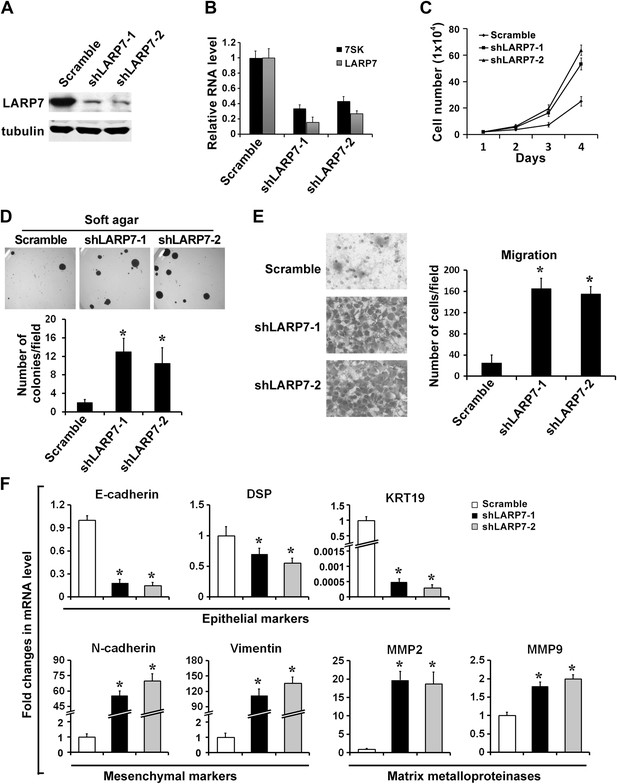
Silencing LARP7 accelerates malignant progression of BT474 breast cancer cells.
(A) Western blotting analysis of LARP7 levels in BT474 control and two shLARP7 pools. (B) qRT-PCR analysis confirms the reduced RNA levels of 7SK and LARP7 in BT474 cells stably expressing shLARP7s. (C–E) Silencing LARP7 in BT474 cells results in increased cell proliferation (C), anchorage-independent growth (D), and cell migration (E). (F) qRT-PCR analysis of various EMT markers and MMPs in control and two shLARP7 pools. PCR values were normalized to that of β-actin. Data are presented as the mean ± SD from three independent measurements.
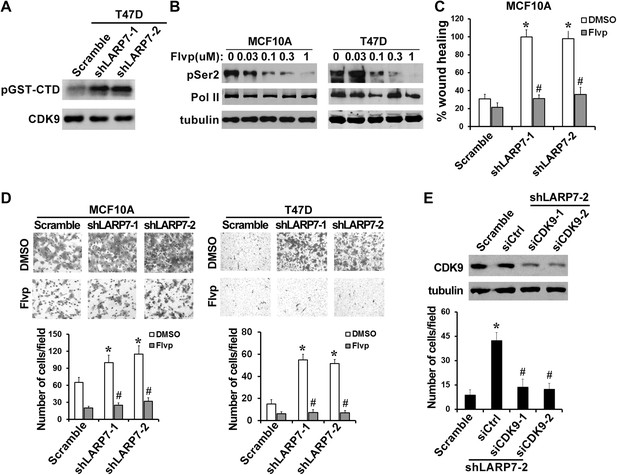
P-TEFb is required for the increased cell migration and invasion induced by LARP7 KD.
(A) An in vitro kinase assay using GST-CTD of Pol II as an exogenous substrate shows that silencing LARP7 results in activation of P-TEFb kinase. (B) Flavopiridol (Flvp) inhibits Ser2 phosphorylation of Pol II CTD in MCF10A and T47D cells. Cells were treated with varying concentrations of flavopiridol for 7 hr before lysis, and pSer2 and total Pol II levels were analyzed by Western blotting. Tubulin was used as a loading control. (C–D) Treatment of cells with 0.3 μM flavopiridol reverses the accelerated cell motility of shLARP7 cells in the wound healing assay (C) and cell migration in the Transwell assay (D). Data are presented as the mean ± SD. p values were determined by the Student's t test. *p<0.05; comparison was between shLARP7 and scramble groups under DMSO treatment; #p<0.05; comparison was between Flvp- and DMSO-treated cells within the same cell lines. (E) Silencing CDK9 by siRNA reverses shLARP7-induced increase in cell migration. Two siCDK9s (siCDK9-1 and siCDK9-2) were transfected into T47D shLARP7-2 cells, and cell migration was assessed by a Transwell assay and quantified in the graph below. The efficiency of CDK9 knockdown was examined by Western blotting (upper panel). p values were determined by the Student's t test. *p<0.05, compared between shLARP7-2 siCtrl and scramble; #p<0.05, compared between shCDK9s and siCtrl.
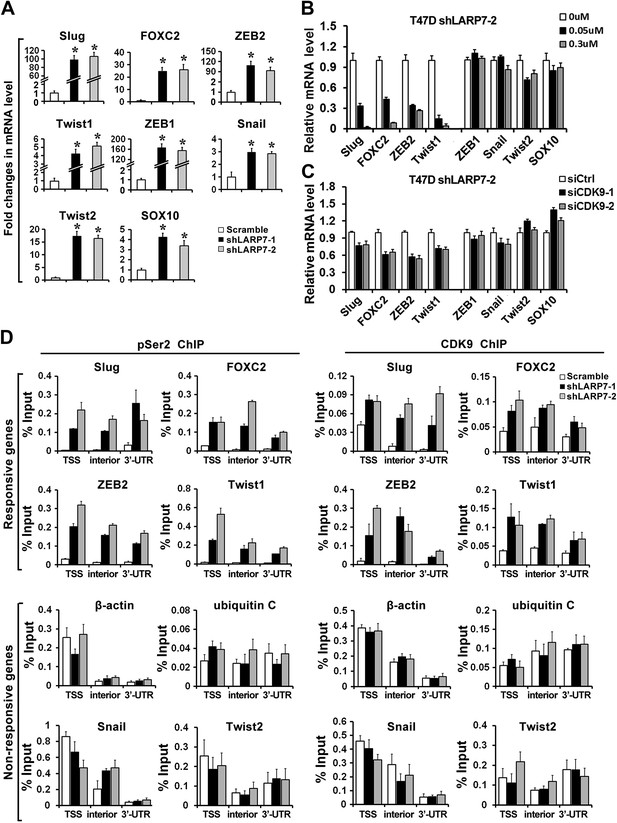
Loss of LARP7 enhances transcription of key EMT genes.
(A) LARP7 knockdown significantly increases the expression of key EMT transcriptional factors in T47D breast cancer cells as measured by qRT-PCR. The p values (*p<0.05) were determined by the Student's t test. (B) Cells were treated with flavopiridol for 7 hr at the indicated concentrations. The expression of EMT genes was assessed by qRT-PCR. (C) Cells were transfected with siCtrl, siCDK9-1 or siCDK9-2. After 48 hr, the expression of EMT genes was assessed by qRT-PCR. (D) Control or two shLARP7 T47D cells were subjected to ChIP analysis to determine the levels of CDK9 and phospho-Ser2 (pSer2) Pol II occupancy at various locations of EMT-related transcription factors and non-responsive genes. qRT-PCR was performed using primers specific to the transcription start site (TSS), interior and 3′-UTR of each gene, and the signals were normalized to that of input. Data are presented as the mean ± SD from three independent measurements.
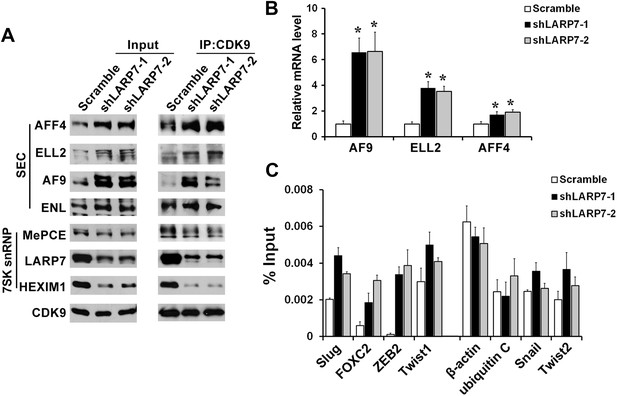
Silencing LARP7 redistributes P-TEFb from the 7SK snRNP to SEC.
(A) Nuclear extracts were prepared from the control or shLARP7 cells and subjected to immunoprecipitation (IP) with anti-CDK9 antibodies. The 7SK snRNP and SEC formation was examined by Western blotting. (B) qRT-PCR analysis of AF9, ELL2, and AFF4 expression in T47D cells with or without LARP7 KD. The p values (*p<0.05) were determined by the Student's t test. (C) ChIP assay was performed to determine the binding of ELL2 to the EMT-related genes and non-responsive genes. Primers at the TSS of each gene were used. Data are presented as the mean ± SD from three independent measurements.
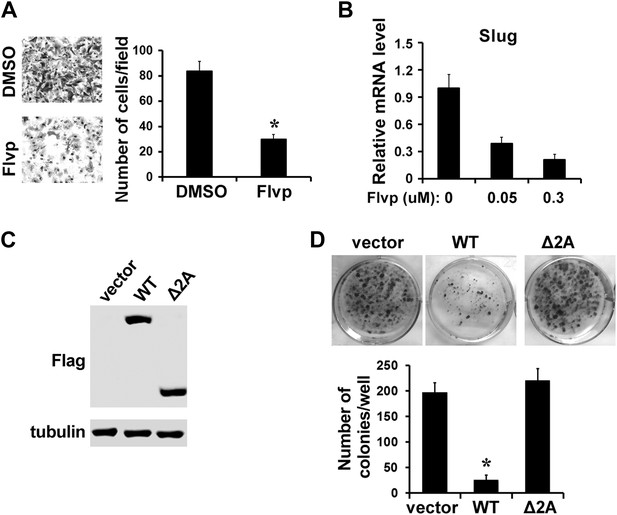
Inhibition of P-TEFb impairs EMT and survival of metastatic MDA-MB-231 cells.
(A) Treatment of MDA-MB-231 cells with 0.3 μM flavopiridol (Flvp) significantly impairs cell migration as shown by the Transwell assay. The p value (*p<0.05) was determined by the Student's t test. (B) qRT-PCR analysis of Slug expression in MDA-MB-231 cells that have been treated with flavopiridol for 7 hr at the indicated concentrations. (C and D) Clonogenic growth assay. MDA-MB-231 cells were transfected with control vector, Flag-tagged WT LARP7, or Δ2A mutant. The expression of LARP7 and Δ2A was confirmed by Western blotting with anti-Flag. (C). (D) Representative culture areas are shown in the top panel. The number of colonies was quantified and shown in the graph below. The p value (*p<0.05) was determined by the Student's t test.
Tables
Association between clinical characteristics and LARP7 levels in breast cancer
| Characteristic | Low levels of LARP7 (n = 148) | High levels of LARP7 (n = 147) | p value |
|---|---|---|---|
| Age | |||
| <45 yr | 95 | 70 | 0.003† |
| ≥45 yr | 53 | 77 | |
| Tumor diameter | |||
| <20 mm | 45 | 66 | 0.012† |
| ≥20 mm | 103 | 81 | |
| ER status | |||
| Negative | 46 | 23 | 0.002* |
| Positive | 102 | 124 | |
| No. of positive nodes | |||
| 0 | 82 | 69 | 0.117 |
| 1–3 | 44 | 61 | |
| ≥4 | 21 | 17 | |
| Differentiation | |||
| High | 31 | 44 | 0.05† |
| Mediate | 48 | 54 | |
| Poor | 69 | 49 | |
| Metastasis | |||
| No | 83 | 111 | 0.001* |
| Yes | 65 | 36 | |
-
*
p≤0.01.
-
†
p≤0.05 were determined by chi-square test.
Additional files
-
Supplementary file 1
List of qRT-PCR primers.
- https://doi.org/10.7554/eLife.02907.013





