A RanGTP-independent mechanism allows ribosomal protein nuclear import for ribosome assembly
Figures
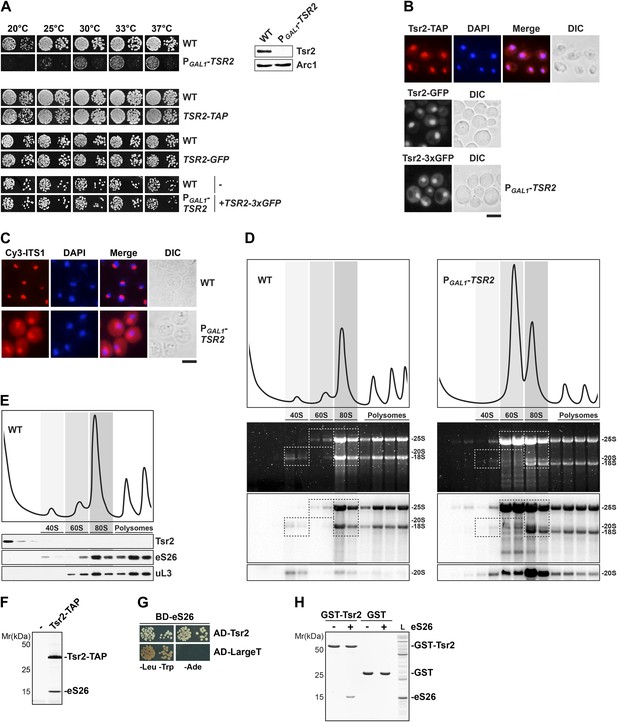
Tsr2 is required for cytoplasmic processing of 20S pre-rRNA to mature 18S rRNA, and directly binds eS26.
(A) Tsr2-TAP, Tsr2-GFP and Tsr2-3xGFP cells are not impaired in growth. Left panel: indicated strains were spotted on glucose containing media in 10-fold dilutions and grown at indicated temperatures for 3–7 days. Right panel: Tsr2 protein levels in whole cell extracts derived from the indicated strains were determined by Western analyses using α-Tsr2 antibodies. Protein levels of Arc1 served as loading control. (B) Tsr2 localizes predominantly to the nucleus. The Tsr2-TAP and the Tsr2-GFP strain and the PGAL1-TSR2 strain containing a centrometric plasmids encoding Tsr2-3xGFP were grown at 30°C to mid-log phase. Localization of Tsr2-TAP was visualized by indirect immunofluorescence microscopy using polyclonal α-TAP antibody (red). Nuclear and mitochondrial DNA was stained with DAPI (blue). Localization of Tsr2-GFP and Tsr2-3xGFP was analyzed by fluorescence microscopy. Scale bar = 5 µm. (C) Tsr2-deficient cells accumulate immature 20S pre-rRNA in the cytoplasm. WT and PGAL1-TSR2 cells were grown at 30°C in glucose containing media to mid-log phase. Localization of 20S pre-rRNA was analyzed by FISH using a Cy3-labeled oligonucleotide complementary to the 5′ portion of ITS1 (red). Nuclear and mitochondrial DNA was stained with DAPI (blue). Scale bar = 5 µm. (D) Tsr2-depleted cells accumulate 80S-like particles. WT and PGAL1-TSR2 cells were grown at 30°C in glucose containing media to mid-log phase. Cell extracts were prepared after cycloheximde treatment to preserve polysomes and subjected to sedimentation centrifugation on 7–50% sucrose gradients. Polysome profiles at OD254nm were recorded and the peaks for 40S and 60S subunits, 80S ribosomes and polysomes are indicated (top panels). The gradients were fractionated and the RNA was extracted, separated on a 2% Agarose gel, stained with GelRed (Biotium, middle panels) and subsequently analyzed by Northern Blotting using probes against indicated rRNAs (bottom panels). Exposure times for phosphoimager screens were 20 min for 25S and 18S rRNA, and 3–4 hr for 20S pre-rRNAs. (E) Tsr2 does not co-sediment with 40S subunits. WT cells were grown at 30°C to mid-log phase, extracts were prepared and fractionated as described in (D). The polysome profile at OD254nm is shown in the upper panel. The peaks for 40S and 60S subunits, 80S ribosomes and polysomes are indicated. The gradient was fractionated, TCA precipitated and the protein content was assessed by Western analyses using the indicated antibodies. (F) Tsr2-TAP co-enriches the r-protein eS26. Tsr2-TAP was isolated by tandem affinity purification and the Calmodulin-eluate was separated by 4–12% gradient SDS-PAGE and analyzed by Silver staining. The indicated proteins were identified by mass spectrometry. (G) Tsr2 interacts with eS26 in a yeast two-hybrid assay. Plasmids encoding the indicated GAL4 DNA-binding domain (BD) and GAL4 activation domain (AD) fusion proteins were transformed into the yeast reporter strain NMY32. Transformants were spotted in 10-fold serial dilutions onto SDC-Leu-Trp (-Leu-Trp) or SDC-Ade (−Ade) and incubated at 30°C for 4 days. Growth on SDC-Ade indicates a strong two-hybrid interaction. The SV40 Large T antigen served as negative control for these analyses. (H) Tsr2 directly binds eS26 in vitro. GST-Tsr2 was immobilized on Glutathione Sepharose before incubation with an E. coli lysate containing recombinant eS26. After incubation, bound proteins were eluted by SDS sample buffer, separated by SDS-PAGE and visualized by Coomassie Blue staining. L = input.
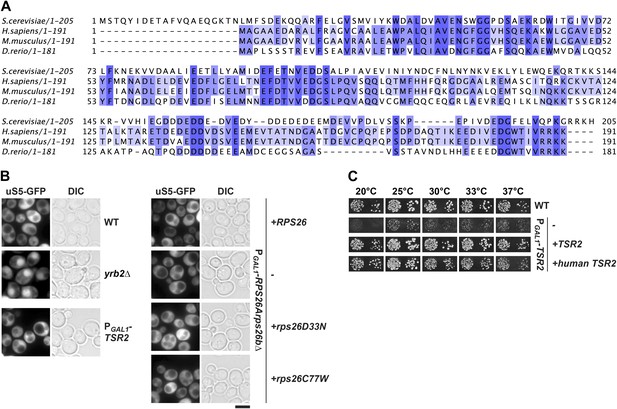
Tsr2 and eS26 depletion does not impair pre-40S nuclear export.
(A) Sequence alignment of Tsr2 from the indicated organisms done by ClustalO (Sievers and Higgins, 2014; Sievers et al., 2011). Conservation at each position is depicted as a gradient from light blue (50% identity) to dark blue (100% identity). (B) Tsr2- and eS26-depletion does not impair pre-40S subunit nuclear export. The indicated strains expressing uS5-GFP were grown in repressive glucose containing liquid media to mid-log phase at 30°C. Localization of uS5-GFP was monitored by fluorescence microscopy. Scale bar = 5 µm. (C) Human Tsr2 rescues the slow growth of Tsr2-depleted cells. The PGAL1-TSR2 cells transformed with indicated plasmids were spotted in 10-fold dilutions on selective glucose containing plates and grown at indicated temperatures for 3–7 days.
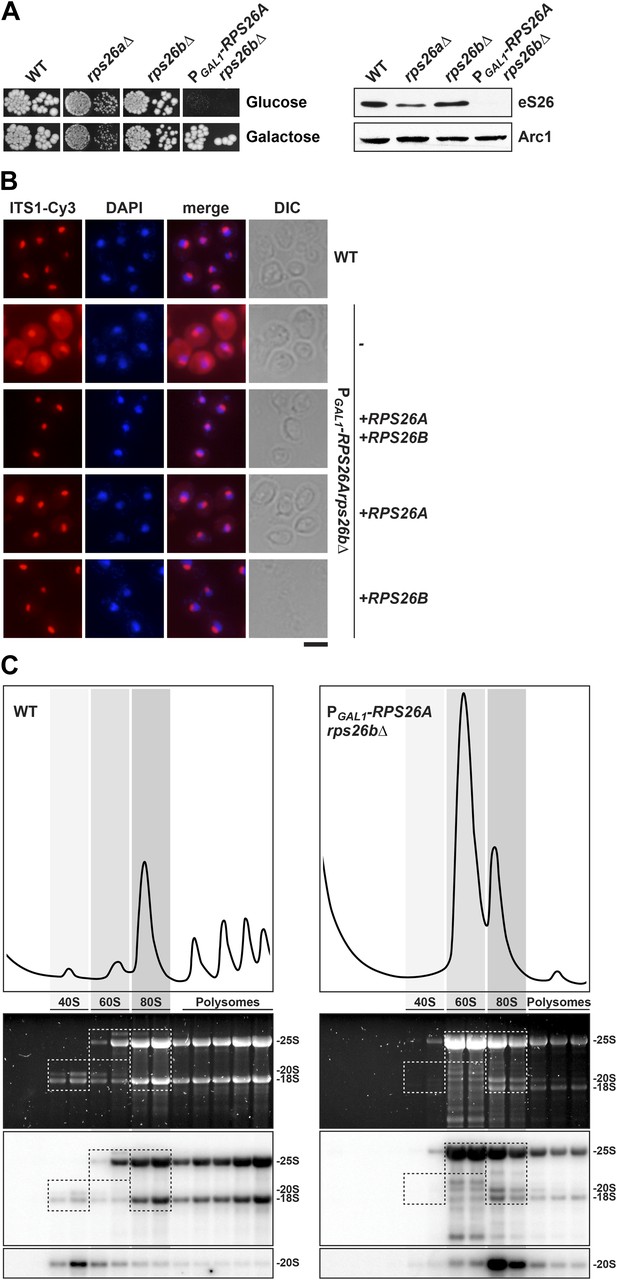
eS26 is required for cytoplasmic processing of immature 20S pre-rRNA to mature 18S rRNA.
(A) eS26 is essential for viability in yeast. Left panel: WT, rps26aΔ, rps26bΔ and the conditional mutant PGAL1-RPS26Arps26bΔ were spotted in 10-fold dilutions on galactose and repressive glucose containing media and grown at 30°C for 2–4 days. Right panel: protein levels of eS26 in whole cell extracts of indicated strains were determined by Western analyses using α-eS26 antibodies. Arc1 protein levels served as loading control. (B) eS26-depleted cells accumulate immature 20S pre-rRNA in the cytoplasm. PGAL1-RPS26Arps26bΔ cells transformed with indicated plasmids were grown in glucose containing liquid media at 37°C to mid-log phase. Localization of 20S pre-rRNA was analyzed by FISH using a Cy3-labeled oligonucleotide complementary to the 5′ portion of ITS1 (red). Nuclear and mitochondrial DNA was stained with DAPI (blue). Scale bar = 5 µm. (C) eS26-depleted cells accumulate 80S-like particles. The indicated strains were grown in glucose containing liquid media at 30°C to mid-log phase. Cell extracts were prepared after cycloheximide treatment and subjected to sedimentation centrifugation on 7–50% sucrose density gradients. Polysome profiles were recorded at OD254nm (top panels). The peaks for 40S and 60S subunits, 80S ribosomes and polysomes are indicated. Sucrose gradients were fractionated, the RNA was extracted, separated on a 2% Agarose gel, stained with GelRed (Biotium, middle panels) and subsequently analyzed by Northern blotting using probes against the indicated rRNAs (bottom panels). Exposure times for phosphoimager screens were 20 min for 25S and 18S rRNA, and 3–4 hr for 20S pre-rRNAs.
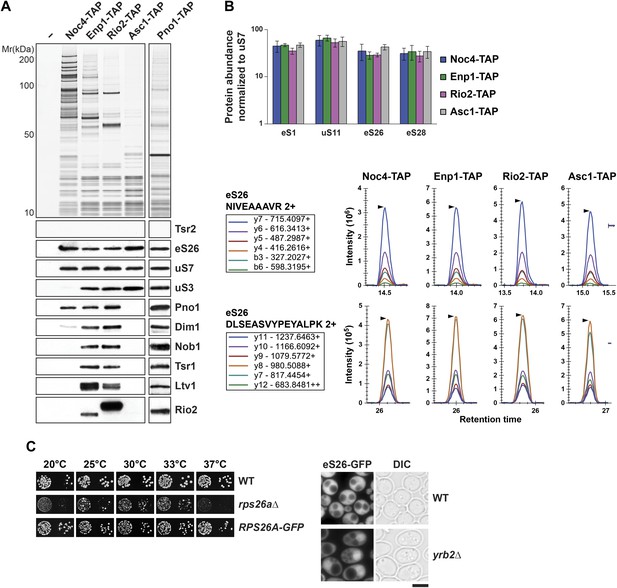
eS26 is incorporated into the earliest pre-ribosome, the 90S.
(A) eS26 co-enriches with pre-ribosomal particles along the 40S maturation pathway. Pre-ribosomal particles in the 40S maturation pathway were purified using the indicated TAP-tagged baits. Calmodulin-eluates were analyzed by Silver staining and Western analyses using the indicated antibodies. The r-protein uS7 served as loading controls for the TAPs. (B) SRM-MS reveals co-enrichment of eS26 with pre-ribosomal particles. Upper panel: the relative abundance of different r-proteins was normalized to uS7 levels in the indicated TAP purifications (three independent biological replicates). The error bars show the standard deviation. Lower panel: the intensity of different transitions (listed in the box) of two specific peptides of eS26 was determined by SRM mass spectrometry in the indicated TAP purifications. (C) eS26-GFP accumulates in the nucleus in a yrb2Δ strain. Left panel: WT, rps26aΔ and RPS26A-GFP cells were spotted in 10-fold dilutions and grown at indicated temperatures for 3–7 days. Right panel: WT and yrb2Δ cells expressing eS26-GFP were grown in glucose containing liquid media to mid-log phase at 20°C. Localization of eS26-GFP was monitored by fluorescence microscopy. Scale bar = 5 µm.
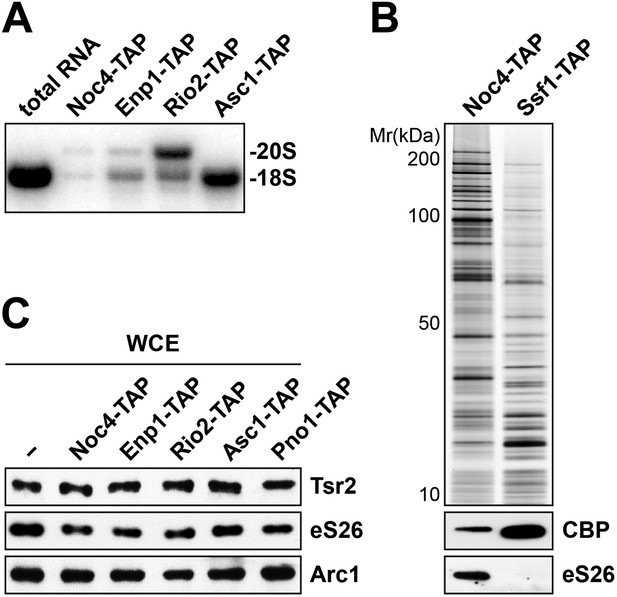
Tsr2 and eS26 protein levels in the indicated TAP strains and levels of 20S pre-rRNA and 18S rRNA in the indicated TAP purified particles.
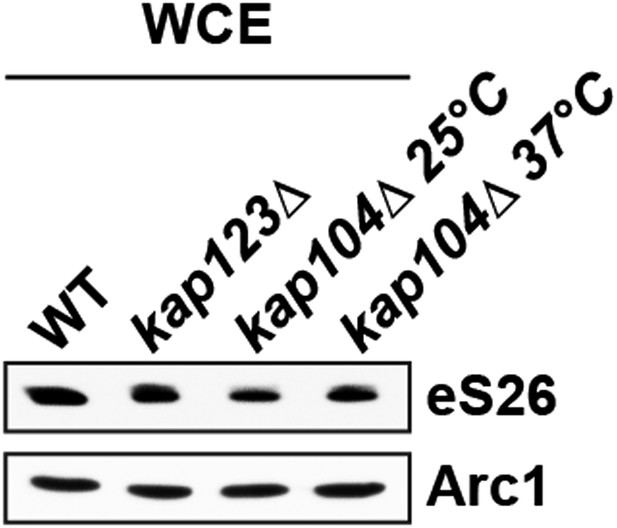
(A) Noc4-, Enp1- and Rio2-TAP purify pre-40S subunits containing immature 20S pre-rRNA whereas Asc1-TAP purifies a 40S subunit containing mature 18S rRNA. 1 µg of RNA isolated from the indicated pre-40S TAP-eluates was separated on a 2% Agarose gel and probed against indicated rRNAs by Northern blotting. 1 µg of total RNA extracted from WT cells was used as a control. (B) eS26 does not co-enrich with the earliest 60S pre-ribosome. Noc4-TAP, the earliest pre-ribosomal particle and Ssf1-TAP, the earliest pre-ribosome in the 60S maturation pathway were isolated. The Calmodulin eluates were visualized by Silver staining and by Western analyses using the indicated antibodies. The CBP signal served as loading controls for the TAPs. (C) Tsr2 and eS26 protein levels in indicated TAP strains (also used in Figure 3A) are equal to levels in WT cells. Whole cell extracts (WCE) were prepared from the indicated strains and analyzed by Western analyses using antibodies against Tsr2 and eS26. The protein Arc1 served as loading control.
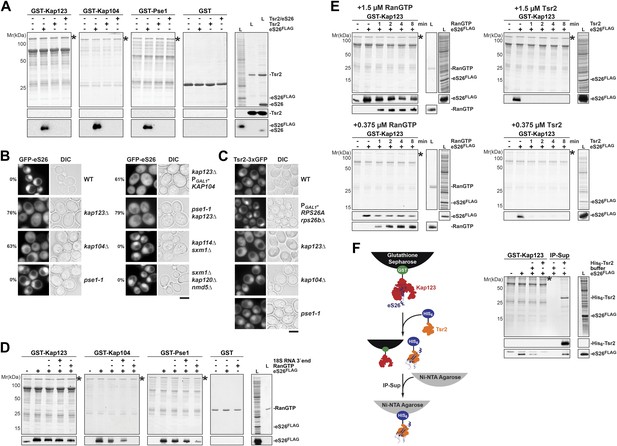
Kap123, Kap104 and Pse1 transport eS26 to the nucleus.
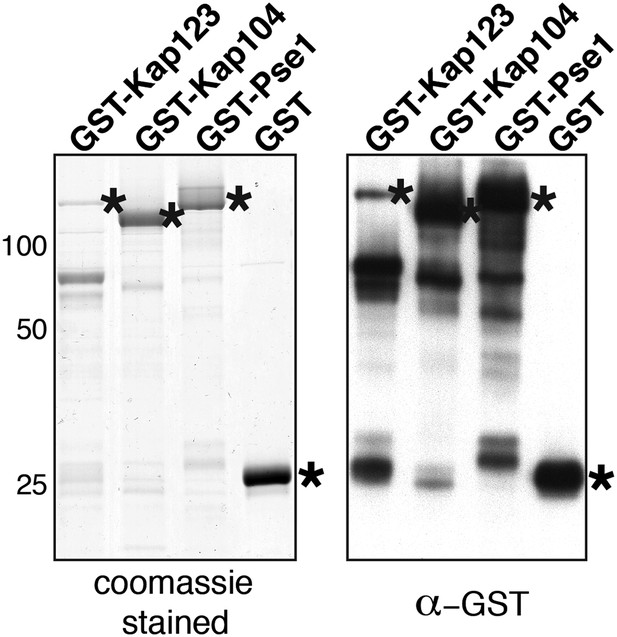
(A) eS26, but not Tsr2:eS26 or Tsr2, interacts with Kap123, Kap104 and Pse1. Recombinant, GST-Kap123, GST-Kap104, GST-Pse1 and GST alone were immobilized on Glutathione Sepharose and incubated with purified 3.4 µM Tsr2, 4 µM Tsr2:eS26, or E. coli lysate containing ∼4 µM eS26FLAG in PBSKMT combined with competing E. coli lysates for 1 hr at 4°C. After washing with PBSKMT, bound proteins were eluted in SDS sample buffer and separated by SDS-PAGE. Proteins were visualized by Coomassie Blue staining or Western analyses using indicated antibodies. L = input. GST-tagged importins are indicated with asterisks. (B) Nuclear uptake of GFP-eS26 is impaired in kap123Δ and kap104Δ mutants. Strains expressing GFP-eS26 were grown in synthetic media at 25°C (ts-mutants: pse1-1 and kap104Δ) or 30°C to mid-log phase. Ts-mutant strains were then shifted to 37°C for 4 hr and localization of GFP-eS26 was analyzed by fluorescence microscopy. Percentage of cells displaying cytoplasmic mislocalization of the GFP-eS26 fusion is indicated. Scale bar = 5 µm. (C) Tsr2-3xGFP is targeted to the nucleus by Kap123. Importin mutant strains expressing Tsr2-3xGFP were grown in synthetic media at 25°C (ts-mutants: pse1-1 and kap104Δ) or 30°C to mid-log phase. Pse1-1 and kap104Δ cells were then shifted to 37°C for 4 hr. PGAL1-RPS26Arps26bΔ cells containing Tsr2-3xGFP were grown for 15 hr in glucose containing media. Localization of Tsr2-3xGFP was analyzed by fluorescence microscopy. Scale bar = 5 µm. (D) RanGTP (His6-Gsp1Q71L-GTP) does not efficiently release eS26 from Kap123 and Pse1. GST-importin:eS26FLAG complexes immobilized on Glutathione Sepharose were incubated with either buffer alone or with 1.5 µM RanGTP or 3 nM 3′-end of 18S rRNA for 1 hr at 4°C. Washing, elution, and visualization were performed as in (A). GST-tagged importins are indicated with asterisks. (E) Tsr2 efficiently dissociates the Kap123:eS26FLAG complex. The GST-Kap123: eS26FLAG complex immobilized on Glutathione Sepharose was incubated with either buffer alone or with 1.5 µM or 375 nM RanGTP or 1.5 µM or 375 nM Tsr2. Samples were withdrawn at the indicated time points. Washing, elution, and visualization were performed as in (A). GST-tagged Kap123 is indicated with an asterisk. (F) eS26 stably associates with Tsr2 after its release from Kap123. Left panel indicates the experimental setup as flowchart. Immobilized GST-Kap123:eS26FLAG complex was incubated with 1.5 µM His6-Tsr2 or buffer alone. As shown in the flowchart, the supernatant was incubated with Ni-NTA Agarose for 1 hr at 4°C (IP-Sup). Washing, elution, and visualization were performed as in (A). GST-tagged Kap123 is indicated with an asterisk.
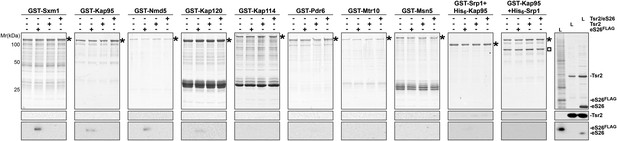
eS26, but not Tsr2:eS26 or Tsr2, interacts with importins.
Recombinant GST tagged importins, immobilized on Glutathione Sepharose, were incubated with purified 3.4 µM Tsr2, 4 µM Tsr2:eS26 or E. coli lysate containing ∼4 µM eS26FLAG in PBSKMT and competing E. coli lysates for 1 hr at 4°C. After washing, bound proteins were eluted in SDS sample buffer, separated by SDS-PAGE, and visualized by either Coomassie Blue staining or Western analyses using indicated antibodies. L = input. GST-tagged importins are indicated with asterisk, His6-Srp1 is indicated with a rectangle.
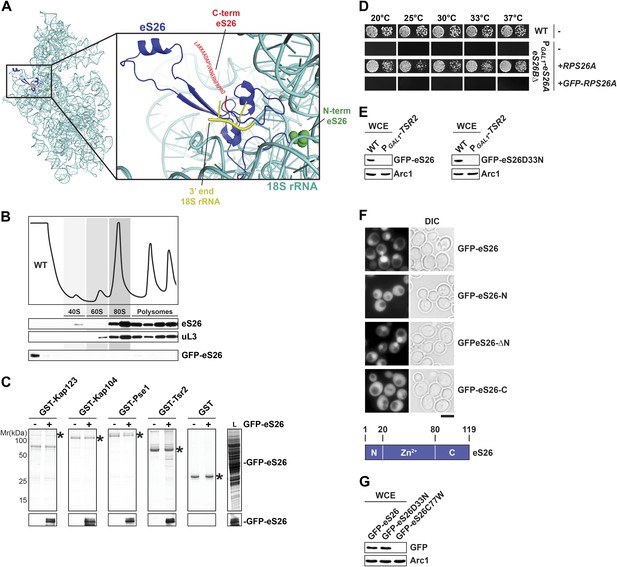
GFP-eS26 binds to importins and Tsr2 but is not incorporated into pre-ribosomes.
(A) Location of N- and C-terminus of eS26 within the mature 40S subunit (Rabl et al., 2011). eS26 N-terminus (green) is embedded deeply within the 40S subunit whereas the C-terminus (red) projects away from the body of the 40S subunit. Red letters indicate the 20 C-terminal residues that are not visualized in the structure (B) GFP-eS26 is not found in heavier fractions on sucrose gradients. WT lysates and lysates containing GFP-eS26 were subjected to sucrose gradient sedimentation as described in Figure 1D. The peaks for 40S and 60S subunits, 80S ribosomes and polysomes are indicated. The proteins in the gradient were detected by Western analyses using the indicated antibodies. (C) GFP-eS26 binds to Kap123, Kap104 and Pse1. Recombinant GST-Kap123, -Kap104, -Pse1 and GST alone were immobilized on Glutathione Sepharose and then incubated with E. coli lysate containing GFP-eS26 for 1 hr at 4°C. Bound proteins were eluted in SDS sample buffer, separated by SDS-PAGE and visualized by Coomassie Blue staining and Western analyses using α-GFP antibody. L = input. (D) GFP-eS26 is unable to rescue the lethality of the eS26 deficient strain. The PGAL1-RPS26Arps26bΔ strain transformed with different plasmids encoding eS26 or GFP-eS26 were spotted in 10-fold dilutions on selective glucose containing plates and grown at indicated temperatures for 3–7 days. (E) GFP-eS26 and GFP-eS26D33N levels are strongly reduced in Tsr2-depleted cells. Whole cell extracts (WCE) prepared from WT and Tsr2-depleted cells were assessed by Western analyses using antibodies against the indicated proteins. Arc1 protein levels served as loading control. (F) Upper panel: the Zn2+-binding domain of eS26 is required for efficient nuclear uptake. WT cells expressing GFP-eS26 truncations were grown in synthetic media at 30°C to mid-log phase and the localization of GFP-eS26 truncations was analyzed by fluorescence microscopy. Scale bar = 5 µm. Lower panel: Schematic for the eS26 truncations used for fluorescence microscopy. (G) GFP-eS26C77W protein levels are strongly reduced in (WCE) extracts. Whole cell extracts were prepared from PGAL1-RPS26Arps26bΔ cells transformed with plasmids encoding for GFP-eS26 WT and mutant proteins. eS26 protein levels were assessed by Western analyses using α-GFP antibodies. Arc1 served as loading control.

Tsr2 efficiently releases the conserved eS26 from importins.
(A) Left panel: sequence alignment of eS26 from the indicated organisms done by ClustalO (Sievers and Higgins, 2014; Sievers et al., 2011). Conservation at each position is depicted as a gradient from light blue (50% identity) to dark blue (100% identity). Mutated residues linked to DBA are depicted with orange (Asp33) and green (Cys77) dots. Right panel: location of eS26 within the mature 40S subunit (Rabl et al., 2011). eS26 clamps the 3′-end of the mature 18S rRNA at the site where the endonuclease Nob1 cleaves the immature 20S pre-rRNA. Inset depicts the 3′-end portion of 18S rRNA (red) in contact with eS26 (blue). The position of amino acids D33 (orange) and C77 (green) that are mutated in DBA or KFS and the coordinated Zn2+ ion (black) are depicted. (B) RanGTP and the 3′-end of 18S rRNA cannot dissociate the Kap123:eS26 complex. GST-Kap123:eS26aFLAG complexes, immobilized on Glutathione Sepharose, were incubated with buffer alone or with 1.5 µM RanGTP, 1.5 µM Tsr2, 3 nM of the 3′-end of 18S rRNA or the combination of RanGTP and the 3′ end of 18S rRNA for 1 hr at 4°C. Bound proteins were eluted in SDS sample buffer, separated by SDS-PAGE and visualized by Coomassie Blue staining and Western analyses using α-eS26 antibodies. L = input. GST-tagged importins are indicated with asterisks. (C) eS26 stably associates with Tsr2 after its release from Pse1. Immobilized GST-Pse1:eS26FLAG complex was treated with 1.5 µM His6-Tsr2 or buffer alone. The supernatant was incubated with Ni-NTA Agarose for 1 hr at 4°C (IP-Sup). Washing, elution, and visualization were performed as in Figure 4E. GST-tagged Pse1 is indicated with an asterisk. (D) RanGTP, but not Tsr2 dissociated the Pse1:Slx9 complex in vitro. Pse1:Slx9 complexes were immobilized on Glutathione Sepharose and incubated with buffer alone or with 1.5 µM RanGTP, 1.5 µM Tsr2 or 3 nM 3′-end of 18S rRNA for 1 hr at 4°C and analyzed as in Figure 4C. GST-tagged importins are indicated with asterisks. (E) Tsr2 efficiently dissociates importin:eS26FLAG complexes. GST-Kap104: eS26FLAG and GST-Pse1:eS26FLAG complexes immobilized on Glutathione Sepharose were incubated with either buffer alone or with 1.5 µM or 375 nM RanGTP or 1.5 µM or 375 nM Tsr2. Samples were withdrawn at the indicated time points (1, 2, 4, 8 min). Washing, elution, and visualization were performed as in Figure 4A. GST-tagged importins are indicated with asterisks.
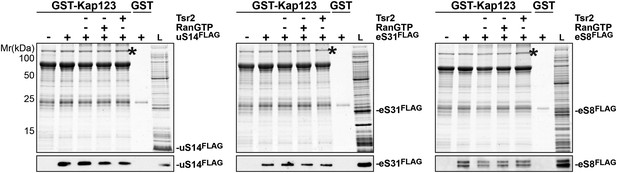
RanGTP and Tsr2 do not release eS31, eS8 and uS14 from Kap123.
GST-Kap123 and GST alone were immobilized on Glutathione Sepharose and incubated with E. coli lysate containing ∼4 µM eS14FLAG, eS31FLAG or eS8FLAG in PBSKMT combined with competing E. coli lysates for 1 hr at 4°C.GST-Kap123:eS14FLAG, GST-Kap123:eS31FLAG, GST-Kap123:eS8FLAG complexes were incubated with either buffer alone or with 1.5 µM RanGTP or 1.5 µM Tsr2 for 1 hr at 4°C. Bound proteins were eluted in SDS sample buffer and separated by SDS-PAGE. Proteins were visualized by Coomassie Blue staining or Western analyses using α-FLAG-antibodies. L = input. GST-Kap123 is indicated with asterisks.
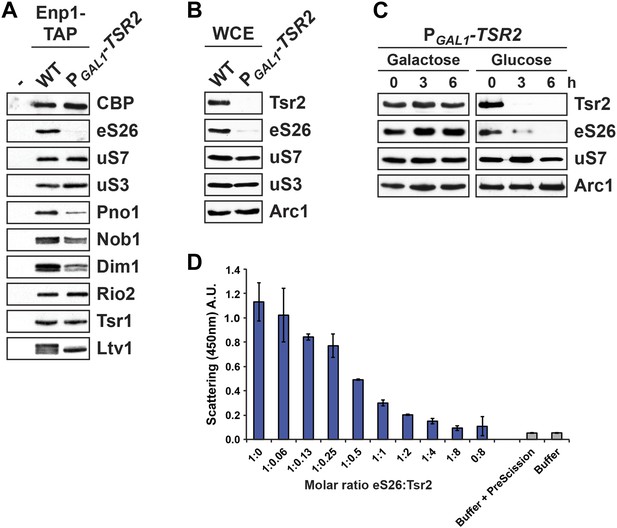
Tsr2 shields eS26 from proteolysis and aggregation, and promotes safe transfer to the 90S pre-ribosome.
(A) Efficient recruitment of eS26 to Enp1-TAP requires Tsr2. Enp1-TAP was isolated from WT and Tsr2-depleted cells. After tandem affinity purification, eluates were separated by 4–12% gradient SDS-PAGE and subjected to Western analyses using indicated antibodies. CBP (α-TAP) levels served as loading control. (B) eS26 levels are strongly reduced in Tsr2-depleted cells. Whole cell extracts (WCE) prepared from WT and Tsr2-depleted cells were assessed by Western analyses using antibodies against the indicated proteins. Arc1 protein levels served as loading control. (C) Tsr2 protects eS26 from proteolysis in vivo. The conditional mutant strain PGAL1-TSR2 growing on galactose medium was transferred to repressive glucose containing liquid media at 30°C. Cells were withdrawn at the indicated time points and whole cell extracts were prepared. Western analyses were performed to determine the levels of the indicated proteins. Arc1 served as loading control. (D) Tsr2 prevents aggregation of recombinant eS26 in vitro. The aggregation assay was performed in a 384-well plate. In each well 33 µM GST-eS26 and a given concentration of Tsr2 (0 up to 266 µM) in PBSKMT was pre-incubated for 1 hr at 4°C (final volume: 90 µl). 250 nM of PreScission protease was added to initiate aggregation. After 1 hr of incubation, the scattering signal of the aggregated eS26 was monitored by a 384-well plate reader by measuring the intensity at 450 nm (Y-axes). Concentration of Tsr2 used in the assay (X-axes) are expressed as a molar ratio of eS26:Tsr2. Four replicates for each well were measured. The error bars show the standard deviation.
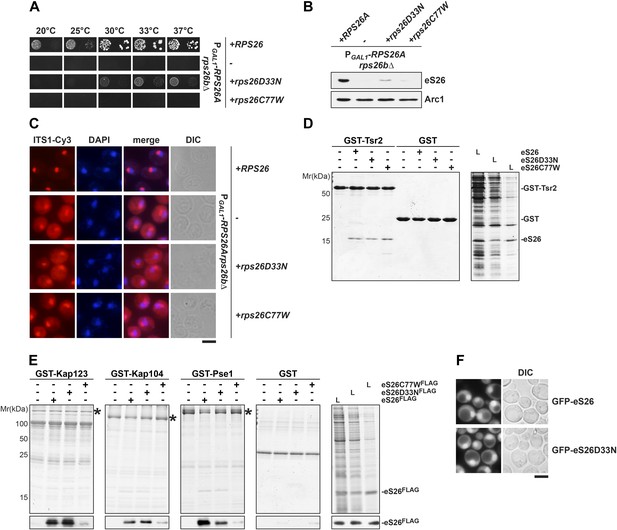
The eS26C77W mutant associated with Klippel-Feil syndrome in Diamond-Blackfan anemia patients is impaired in binding importins.
(A) The DBA linked eS26D33N and eS26C77W mutants are unable to fully rescue the growth defect of eS26-depleted cells. The PGAL1-RPS26Arps26bΔ strain transformed with different plasmids encoding eS26 mutants were spotted in 10-fold dilutions on selective glucose containing plates and grown at indicated temperatures for 3–7 days. Residues mutated in DBA are depicted in Figure 4—figure supplement 3A. (B) DBA linked mutations cause strongly reduced eS26 protein levels. Whole cell extracts were prepared from PGAL1-RPS26Arps26bΔ cells transformed with indicated plasmids encoding for eS26 WT and mutant proteins. eS26 protein levels were assessed by Western analyses using α-eS26 antibodies. Arc1 served as loading control. (C) eS26 mutants linked to DBA accumulate 20S pre-rRNA in the cytoplasm. PGAL1-RPS26Arps26bΔ cells transformed with plasmids encoding for eS26 WT and mutant proteins were grown at 37°C to mid-log phase in glucose containing medium. Localization of 20S pre-rRNA was analyzed by FISH using a Cy3-labeled oligonucleotide complementary to the 5′ portion of ITS1 (red). Nuclear and mitochondrial DNA was stained with DAPI (blue). Scale bar = 5 µm. (D) Tsr2 interacts with eS26 mutants linked to DBA. Recombinant GST-Tsr2 was immobilized on Glutathione Sepharose and then incubated with E. coli lysates containing eS26aFLAG, eS26D33NFLAG or eS26C77WFLAG lysates for 1 hr at 4°C. Bound proteins were eluted by SDS sample buffer, separated by SDS-PAGE and detected by Coomassie Blue staining. L = input. (E) eS26C77W is impaired in binding to Kap123, Kap104 and Pse1. Recombinant GST-Kap123, -Kap104, -Pse1 and GST alone were immobilized on Glutathione Sepharose and then incubated with E. coli lysate containing eS26FLAG, eS26D33NFLAG or eS26C77WFLAG for 1 hr at 4°C. Bound proteins were eluted in SDS sample buffer, separated by SDS-PAGE and visualized by Coomassie Blue staining and Western analyses using α-eS26 antibody. L = input. (F) The GFP-eS26D33N fusion protein is efficiently targeted to the nucleus. WT cells expressing GFP-eS26 and GFP-eS26D33N were grown in synthetic media at 30°C to mid-log phase and the localization of GFP-eS26 was analyzed by fluorescence microscopy. Scale bar = 5 µm.
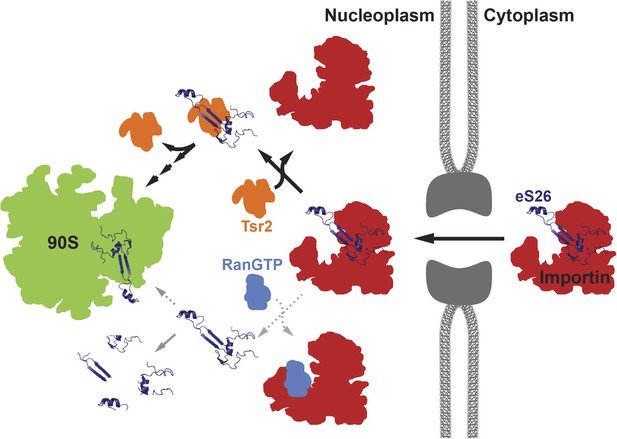
A model for the transport of eS26 to the 90S pre-ribosome.
Newly synthesized eS26 is transported from the cytoplasm into the nucleus by importins. In the nucleus, Tsr2 alone removes eS26 from importins by a RanGTP-independent mechanism. Subsequently, Tsr2 binds the released eS26, protects it from proteolysis and aggregation, and enables safe transfer to the 90S pre-ribosome. If eS26 is released from the importin by RanGTP it may not immediately encounter Tsr2, resulting in a smaller fraction reaching the 90S pre-ribosome. See ‘Discussion’ for details of the proposed model.
Additional files
-
Supplementary file 1
(A) Yeast strains used in this study. (B) Plasmids used in this study.
- https://doi.org/10.7554/eLife.03473.016
-
Supplementary file 2
Peptides used for SRM assays.
- https://doi.org/10.7554/eLife.03473.017