Pvr expression regulators in equilibrium signal control and maintenance of Drosophila blood progenitors
Figures
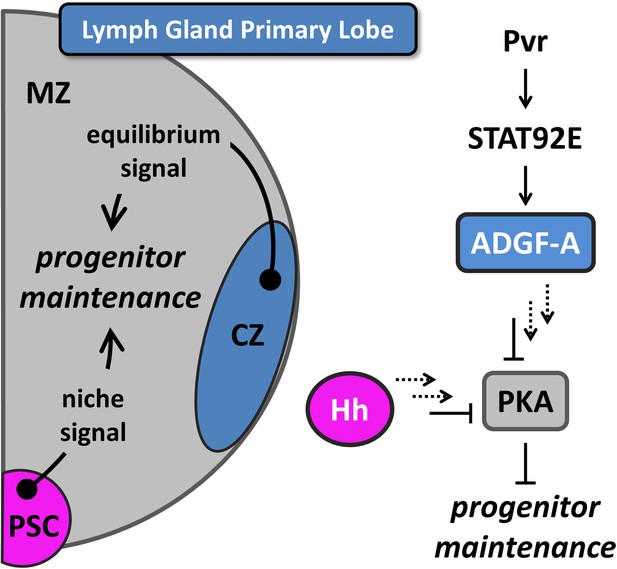
Equilibrium signaling maintains hematopoietic progenitors in the developing lymph gland.
The lymph gland primary lobe consists of three distinct cellular populations or zones. The medullary zone (MZ) contains blood progenitor cells while the nearby cortical zone (CZ) contains differentiating and mature blood cells. The posterior signaling center (PSC) functions as a supportive population (a niche) that expresses Hedgehog (Hh) and maintains the progenitor cells utilizing this ‘niche signal’. The receptor tyrosine kinase (RTK) Pvr and the STAT (STAT92E) transcriptional activator are required in CZ cells for the proper expression and secretion of the extracellular enzyme ADGF-A, which keeps the extracellular adenosine levels relatively low by converting it to inosine. The Pvr ligand Pvf1 is made in PSC cells and is transported through the lymph gland to activate Pvr in CZ cells. Collectively, we refer to the system that generates ADGF-A from the differentiating cells as ‘equilibrium signaling’, which is required independently of the niche-derived Hh signaling for the maintenance of progenitor blood cells in the MZ. Signaling events downstream of both ADGF-A and Hh (dashed arrows) cause the inhibition of Protein Kinase A (PKA) within progenitor blood cells, thereby promoting their maintenance. The individual components are color coded to match the schematic of the lymph gland. The equilibrium signal ADGF-A is blue, originating from the CZ; the niche signal Hh is magenta, originating in the PSC; PKA is gray, functioning in the MZ progenitor cells. Full details of this molecular pathway can be found in Mondal et al., (2011).

The Hand-Hemolectin Lineage Tracing-gal4 line (HHLT-gal4 UAS-2XEGFP) and its use as an in vivo screening tool.
(A) Schematic describing the key elements of the HHLT-gal4 driver line. (B) Image showing the hematopoietic system within a wandering stage third-instar HHLT > GFP larva (dorsal view). Primary, secondary, and tertiary lobes of the lymph gland are readily discernible through overlying musculature, epidermal cells, and cuticle. Lymph gland lobes develop bilaterally, flanking the larval heart (dorsal vessel, DV). Non-blood pericardial cells (PC) also express GFP due to early expression of Hand-gal4. Circulating/sessile blood cells also express GFP due to Hml-gal4 and sessile groups are easily observable. GFP is also seen in ventrally located salivary glands (SG, out of focus) of larvae beyond the third-instar transition (due to Hand-gal4). (C) HHLT > GFP control larvae; (D) HHLT > GFP larvae overexpressing Ras85D (LA 527) exhibit hyperproliferative lymph glands; (E) HHLT > GFP larvae overexpressing combgap (LA 630) show little or no GFP expression in the lymph gland region. Arrows indicate GFP fluorescence from salivary glands (SG).
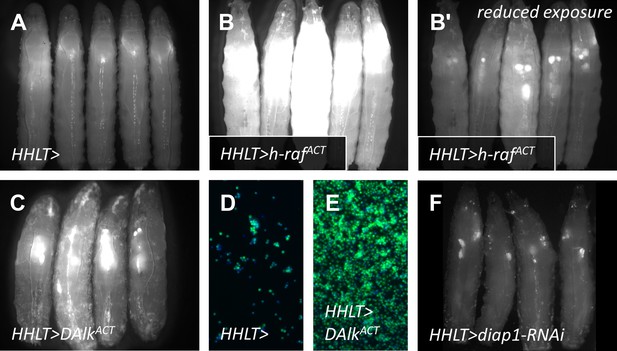
As a ‘proof-of-principle’ approach and to assess the effectiveness of HHLT-gal4 as a screening tool, HHLT-gal4 was crossed to lines harboring gain-of-function UAS transgenes known to cause excessive cellular proliferation, with the expectation that such transgenes would cause significant expansion of the hematopoietic tissues.
(A) HHLT > GFP larvae (outcrossed to w1118) showing baseline fluorescence. (B–B′) HHLT > GFP larvae expressing UAS-human activated Raf (h-RafACT) show increased GFP fluorescence with the same exposure (B) as for control larvae (A) and exhibit enlarged lymph glands with reduced exposure time (B′). (C) HHLT > GFP larvae expressing UAS-activated Drosophila Alk (DAlkACT) also exhibit enlarged lymph glands. (D and E) Relative bleed cell densities from control animals (D) and animals expressing UAS-DAlkACT (E) which show an expansion. (F) HHLT > GFP larvae expressing UAS-diap1-RNAi exhibit reduced fluorescence in lymph gland and circulating cells. The obvious changes in the level of HHLT-gal4-mediated EGFP expression observed in these backgrounds shows that HHLT-gal4 is indeed a useful tool with which to assess the hematopoietic system in vivo, in the context of a genetic screen.
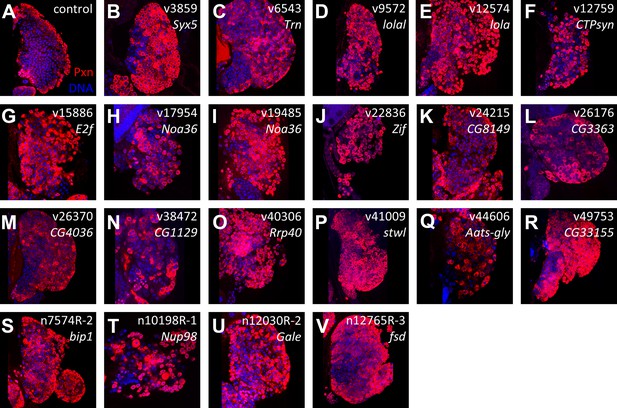
Identification of RNAi lines that cause an expanded Peroxidasin phenotype when expressed throughout the lymph gland.
Peroxidasin (Pxn, red) is normally restricted to cortical zone cells (near the periphery) (A, control) but is seen throughout the lymph gland in RNAi backgrounds (B–V) expressed by HHLT-gal4. Line identifiers and gene targets are shown; additional details listed in Table 1. Images represent a single middle confocal section taken from a Z-plane series through the entire primary lobe.
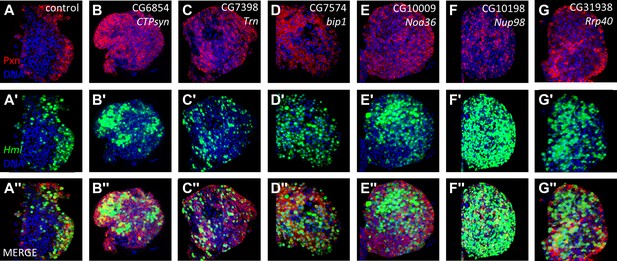
Identification of candidate genes that cause an expanded Peroxidasin expression phenotype within the lymph gland when knocked down by RNAi in differentiating and mature cells.
RNAi from identified lines (Figure 3/Table 1) was expressed in lymph glands using Hml-gal4 UAS-GFP (Hml > GFP). In the control, Pxn (A) and GFP (A′) are restricted to the cortical zone (periphery). By contrast, knock down of six candidate genes causes extensive expression of Pxn (B–G) and Hml (Hml > GFP) (B′–G′) throughout the lymph gland, indicating a loss of progenitors in these genetic backgrounds. The combined Pxn and Hml expression patterns for each genetic background are shown (MERGE, A″–G″). DNA (blue) is stained to mark nuclei.
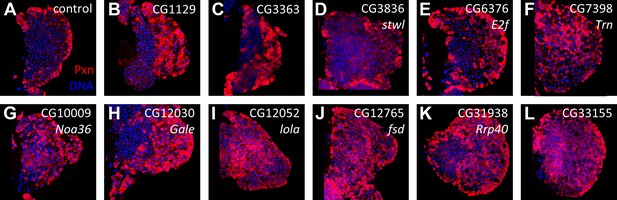
RNAi lines causing an expanded Peroxidasin expression phenotype when expressed in progenitor cells using dome-gal4.
(A) Control (dome-gal4 with no UAS-dsRNA) with Pxn expression (red) limited to the cortical zone of the lymph gland primary lobe; DNA (blue). (B–L) Individual candidate genes identified by RNAi knockdown directly in progenitor cells using dome-gal4. RNAi for each gene causes the expansion of Pxn expression (red) throughout the primary lobes.
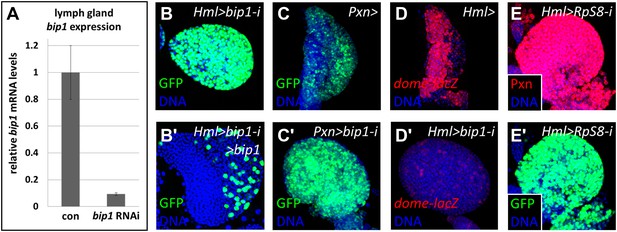
Validation of the bip1 RNAi phenotype.
(A) Quantitative RT-PCR demonstrates that bip1 is expressed in the lymph gland and that the RNAi line NIG 7574R-1 targeting bip1 indeed reduces bip1 transcript level when expressed using Hml-gal4. Hml-gal4 expresses GFP throughout the primary lobes in bip1 RNAi lymph glands (Hml > bip1-i, B), and this phenotype is suppressed by overexpression of bip1 (B′), restoring both the cortical and the medullary zones. (C–C′) Expression of bip1 RNAi using Pxn-gal4 phenocopies obtained with Hml-gal4, further supporting a cell-type-specific function of bip1. Expression of the progenitor cell marker dome-MESO-lacZ (D) is strongly reduced in bip1 RNAi lymph glands (D′), demonstrating that the gain in differentiation markers is due to the loss of progenitor cells that normally express dome-MESO-lacZ. RNAi knock down of RpS8, encoding a putative Bip1-interacting protein, causes the expansion of Pxn and Hml-gal4 expression throughout the lymph gland (E–E′), similar to that observed upon the loss of bip1.
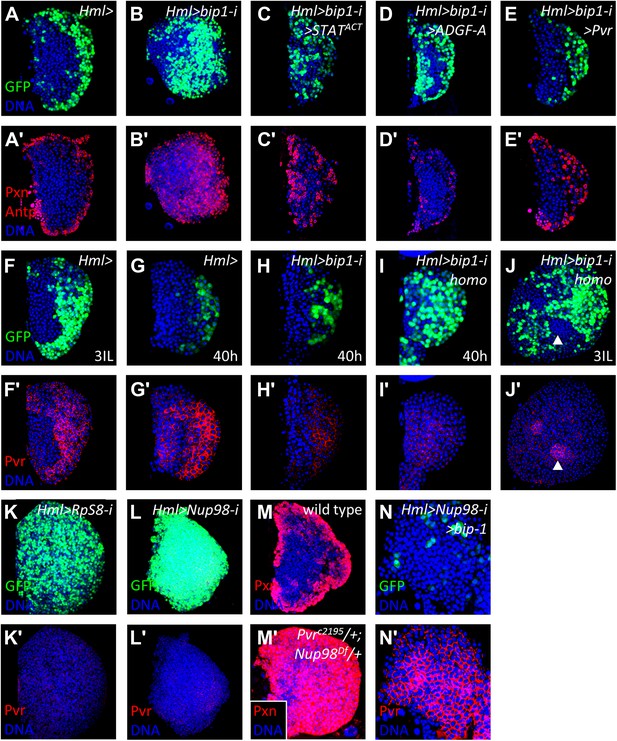
bip1, RpS8, and Nup98 control Pvr expression in the lymph gland.
Expression of bip1 RNAi in differentiating cells (Hml-gal4 or Hml>, B–B′) causes expansion of both Pxn and Hml-gal4 UAS-GFP throughout the lymph gland, as compared to controls (A–A′). Misexpression of either activated STAT (STATACT, C–C′), ADGF-A (D–D′), or Pvr (E–E′) partially (in the case of STAT activation or ADGF-A overexpression) or fully (in case of Pvr overexpression) suppresses this bip1 phenotype, suggesting that bip1 functions upstream of these genes. Expression of Pvr in control third-instar lymph glands (F–F′) and mid-second instar (40 hr post-hatching, G–G′). Reduced expression of Pvr is already apparent in bip1 RNAi lymph glands by 40 hr (H–H′), and this loss is even stronger in homozygous animals expressing higher levels of RNAi (I–I′); increased differentiation, based upon Hml-gal4 UAS-GFP expression, is also apparent (I). Strong suppression of Pvr is also observed in homozygous bip1 RNAi lymph glands (J–J′). RNAi knockdown of RpS8 also causes differentiation and the loss of Pvr expression (K–K′). Likewise, RNAi knockdown of Nup98 also causes differentiation and the loss of Pvr expression (L–L′). (M) Control background (Hml-gal4/+) showing normal expression of the differentiation marker Pxn in the cortical zone of the lymph gland. Progenitor cells in the MZ region are easily discerned by their lack of Pxn expression. By contrast, few progenitor cells (Pxn-negative cells) are observed in lymph glands when single-copy loss-of-function mutations of Pvr and Nup98 (PvrC2195/+; Nup98Df(3R)mbc-R1/+) are combined (M′), further indicating the close interaction between these genes. The middle-third (confocal z-stack) of the primary lobe is shown. Misexpression of bip1 in this background is sufficient to suppress these phenotypes and restore Pvr expression to the lymph gland (N–N′).
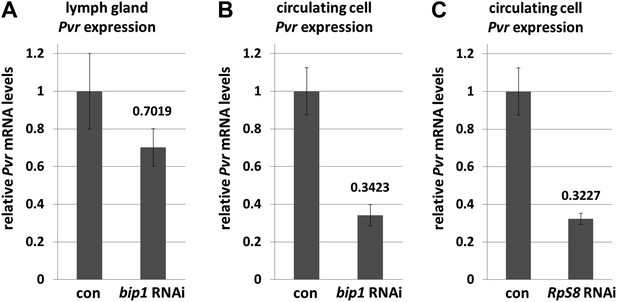
bip1 and RpS8 are required for normal Pvr transcript levels.
Quantitative RT-PCR analysis demonstrating that RNAi knockdown of bip1 in lymph glands (A) and circulating cells (B) using Hml-gal4 causes a reduction in Pvr transcript levels to approximately 70% and 34% of the control (Hml-gal4 only) level, respectively. RpS8 RNAi using Hml-gal4 reduces Pvr to approximately 32% of the control level (C, circulating cells). Note that Hml-gal4 is only expressed in a subset of cells in the lymph gland but in the vast majority of cells in circulation.
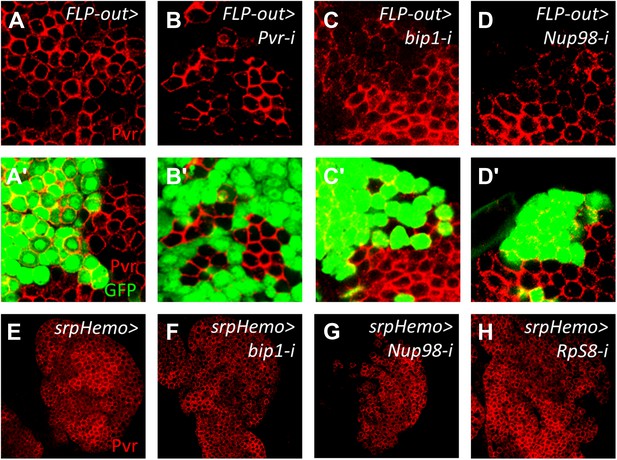
Pvr expression is regulated autonomously by bip1, Nup98, and RpS8 within the lymph gland.
Mock FLP-out Gal4-expressing clones (GFP-positive cells, green) show no reduction in Pvr expression (red, A–A′), whereas similar clones expressing Pvr RNAi show a very strong reduction in Pvr protein expression (B–B′). FLP-out Gal4-expressing clones expressing either bip1 RNAi (C–C′) or Nup98 RNAi (D–D′) also reduce Pvr protein levels, with bip1 RNAi exhibiting somewhat stronger effects. FLP-out clones were made using Hand-gal4 UAS-FLP Ay-gal4 UAS-GFP at 25°C. Restricting the RNAi knockdown of bip1 (F), Nup98 (G), or RpS8 (H) to circulating cells with srpHemo-gal4 (E) shows no effect on lymph gland Pvr levels.
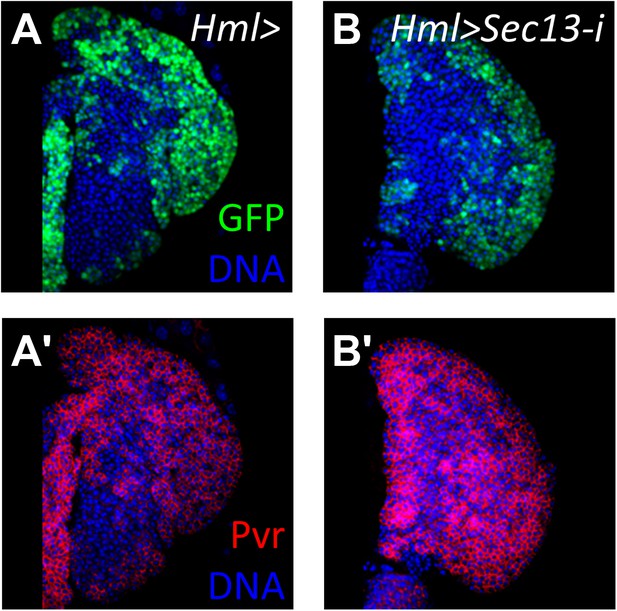
Loss of the nucleoporin Sec13 by RNAi neither causes a differentiation phenotype within the lymph gland nor the loss of Pvr expression.
(A) Hml-gal4 control showing normal cortical zone expression (UAS-GFP). (A′) Pvr is expressed normally throughout the primary lobe in control lymph gland in (A). (B–B′) Loss of Sec13 by RNAi (Hml-gal4 UAS-Sec13-i) does not cause increased differentiation (GFP expression) or the loss of Pvr expression (red). DNA, blue.
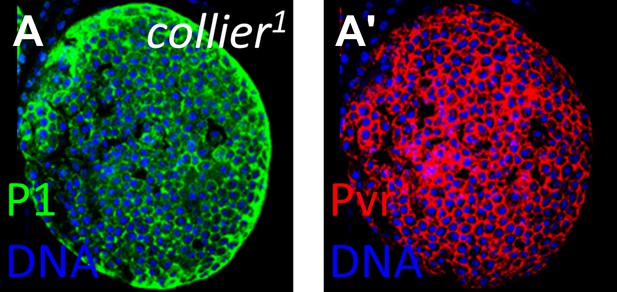
Loss of Pvr expression is not a common feature of highly differentiated lymph glands.
The lymph gland marker P1 is normally restricted to the differentiated cells of the cortical zone. However, in collier1 (col1) mutants that lack niche signaling, P1 expression (A, green) is observed throughout, indicating strong differentiation and a lack of progenitor cells. In this background, Pvr is expressed at normal levels (A′, compare with cortical zone levels in Figure 6F′), indicating that loss of Pvr is not a general feature of highly differentiated lymph glands.
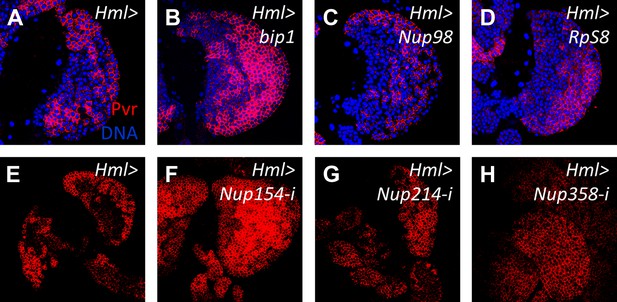
Overexpression of bip1, Nup98, and RpS8, and RNAi knockdown of other nucleoporins does not affect Pvr levels in the lymph gland.
Compared with control levels (A, Hml-gal4), overexpression of bip1 (B, UAS-bip1LA645), Nup98 (C, UAS-Nup98 [Parrott et al., 2011]), or RpS8 (D, UAS-RpS8DP01446 [Staudt t al., 2005]) does not significantly affect Pvr levels (red). Likewise, RNAi knockdown of Nup154 (F), Nup214 (G), or Nup358 (H) (all with Hml-gal4) does not significantly alter Pvr level compared to controls (E), further supporting the specific function of Nup98 in Pvr regulation.
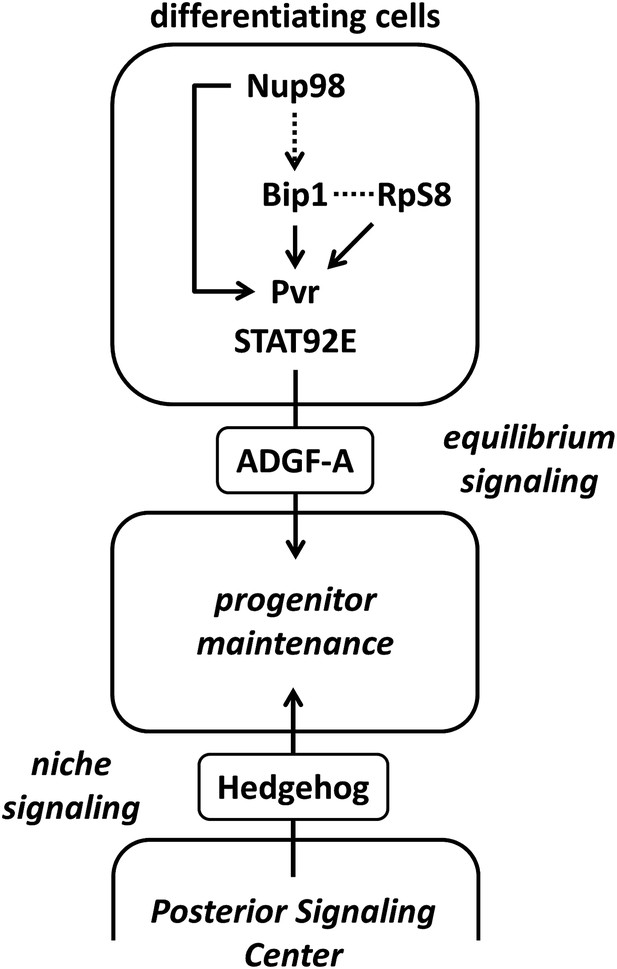
Schematic of the equilibrium signaling pathway demonstrating the proposed roles of Bip1, RpS8, and Nup98 in controlling Pvr.
Bip1, RpS8, and Nup98 are independently required for the expression of Pvr (direct arrows). Rescue of endogenous Pvr expression by misexpression of bip1 in the Nup98 RNAi background indicates that bip1 functions genetically downstream of Nup98 (dashed arrow) in the control of Pvr expression. Bip1 and RpS8 may work together in a complex (dashed line) to control Pvr expression in vivo. These components collectively comprise the known equilibrium signaling pathway working within the lymph gland to promote progenitor cell maintenance, along with the previously known Hh niche signaling mechanism.
Tables
RNAi lines and target genes causing an ‘expanded’ Peroxidasin expression phenotype with HHLT-gal4
| Line # | UAS-RNAi ID | RNAi target | Gene | Off targets | LG size/quality | Protein function |
|---|---|---|---|---|---|---|
| 1 | 3859 | CG4214 | Syx5 | 0 | Small/missing | Golgi SNARE |
| 2 | 6543 | CG7398 | Trn | 1 | Large/baggy | hnRNP nuclear import |
| 3 | 9572 | CG5738 | lolal | 0 | Small | Transcription factor |
| 4 | 12574 | CG12052 | lola | 0 | Large/baggy | Transcription factor |
| 5 | 12759 | CG6854 | CTPsyn | 0 | Small/baggy | CTP synthase |
| 6 | 15886 | CG6376 | E2f | 1 | Small/normal | Transcription factor |
| 7 | 17954 | CG10009 | Noa36 | 0 | Small/missing | Zinc finger nucleolar protein |
| 8 | 19485 | CG10009 | Noa36 | 0 | Small/missing | Zinc finger nucleolar protein |
| 9 | 22836 | CG10267 | Zif | 0 | Small/missing | Transcription factor |
| 10 | 24215 | CG8149 | CG8149 | 0 | Baggy | DNA binding protein |
| 11 | 26176 | CG3363 | CG3363 | 0 | Small | Unknown |
| 12 | 26370 | CG4036 | CG4036 | 1 | Large | Oxidoreductase |
| 13 | 38472 | CG1129 | CG1129 | 0 | Small/missing | Peptide transferase |
| 14 | 40306 | CG31938 | Rrp40 | 0 | Small/normal | RNA exosome |
| 15 | 41009 | CG3836 | stwl | 0 | Small/normal | Transcription factor |
| 16 | 44606 | CG6778 | Aats-gly | 0 | Small/missing | Glycyl-tRNA synthetase |
| 17 | 49753* | CG33155 | CG33155 | 4 | Small/normal | Unknown |
| 18 | 7574R-2 | CG7574 | bip1 | 0 | Small/baggy | Transcription factor |
| 19 | 10198R-1 | CG10198 | Nup98-96 | 0 | Small/missing | Nucleoporin |
| 20 | 12030R-2 | CG12030 | Gale | 0 | Small | UDP-galactose 4'-epimerase |
| 21 | 12765R-3 | CG12765 | fsd | 0 | Small/normal | F-box protein |
-
*
This RNAi line targeting sequence overlaps with the putative mRpL53 gene in the same locus. Lines 1–17 from VDRC, lines 18–21 from NIG Japan.
Additional files
-
Supplementary file 1
Genes and phenotypes associated with P{Mae-UAS.6.11} LA insertion lines expressed with HHLT-gal4. Screen and line identifiers are shown along with the predicted misexpressed gene and the associated whole-animal lymph gland and circulating cell phenotype.
- https://doi.org/10.7554/eLife.03626.018
-
Supplementary file 2
Genes and phenotypes associated with UAS-RNAi lines expressed with HHLT-gal4. Screen and line identifiers are shown along with the targeted gene for knockdown by RNAi, the associated whole-animal lymph gland phenotype based upon GFP expression, and the Peroxidasin (Pxn) expression phenotype of dissected lymph glands.
- https://doi.org/10.7554/eLife.03626.019





