LINKIN, a new transmembrane protein necessary for cell adhesion
Figures
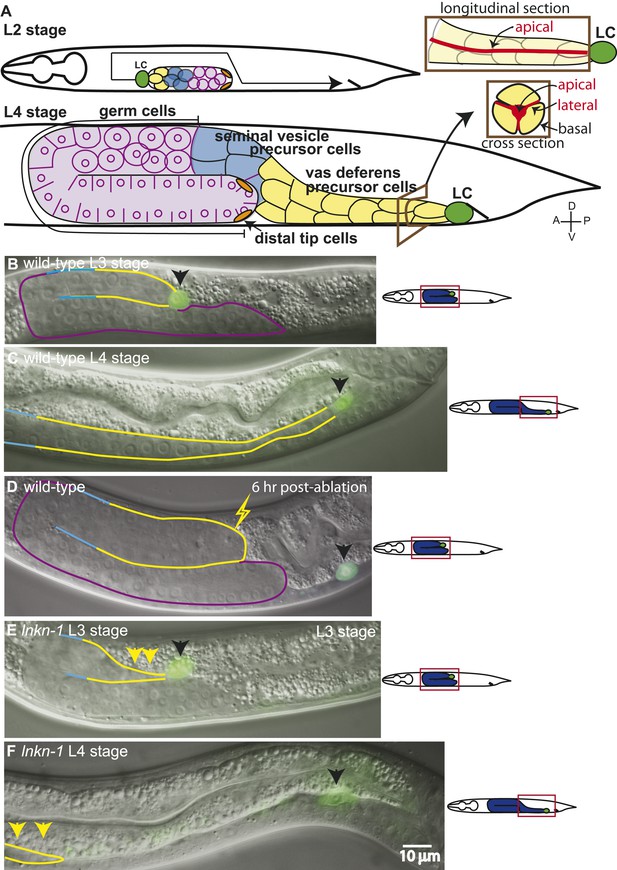
The collective migration of the male gonad in wild-type animals and its disruption in lnkn-1 mutants.
(A) Intact wild-type male gonad shape is generated by the collective migration of somatic and germ cells. The migration of the leader cell, the linker cell (LC; green), pulls the interconnected follower cells. The somatic gonad consists of the LC, the vas deferens precursors (yellow), the seminal vesicle precursors (blue), and the distal tip cells (orange). The germ cells (purple) follow behind most of the somatic gonad. At the beginning of its migration in the early L2 stage, the gonad is a small cluster of cells in the ventral mid-body of the animal (top left panel). As the LC migration defines the shape of the mature gonad, the gonad expands through the proliferation of the interconnected follower cells (bottom left panel). Longitudinal and transverse sections of the vas deferens precursor cells reveal the apical domain (red) running through the somatic gonad core (right panels). (B–F) Nomarski micrographs of gonads superimposed with fluorescence images of YFP-tagged LC (green fluorescence, black arrow). Gonad is outlined in the same color scheme as (A). (B, C) Wld-type L3 and L4 stage gonads. (D) The connection between the LC and the gonad was severed by ablating cells immediately behind the LC and examined 6 hr later. The LC alone has continued to migrate along its normal path, while the gonad no longer elongates after being severed from the LC. (E) In the L3 stage lnkn-1(gk367) mutant, the gonad starts to show thinning of follower cells (yellow arrows) behind the LC (black arrow). (F) By the mid-L4 stage, gonad (yellow arrows) has stopped migrating at the point where the LC detached, while the LC (black arrow) has continued to migrate. In this and subsequent figures, anterior (A) is to the left, posterior (P) is to the right, dorsal (D) is to the top, and ventral (V) is to the bottom.

LINKIN protein domains and sequence are conserved across diverse metazoan species.
(A) C. elegans LNKN-1 is a single-pass transmembrane protein of 599 amino acids (AA). Conserved protein motifs include seven atypical FG–GAP domains (orange boxes) and an extracellular region proximal to the transmembrane domain (light blue box). The gk367 genomic lesion results in the deletion of 92 AAs based on cDNA sequencing. (B) LINKIN sequence from divergent animals (Homo sapiens, Mus musculus, Drosophila melanogaster, and Caenorhabditis elegans) were aligned using Clustalw. The intracellular domain shows high conservation in all examined species. ‘*’ indicates identical AA, ‘:’ indicates strong similarity, and ‘.’ indicates weak similarity. Signal peptide is boxed in purple, extracellular domain in blue, transmembrane domain in gray, and intracellular domain in green. Sequence deleted by gk367 mutation is underlined in red. FG–GAP domains are highlighted in orange, and the Dx(D/N)xDxxxD calcium-binding motif contained within each FG–GAP domain is indicated with red letters. FG–GAP domains were defined as a region from 8 AA N-terminal of the calcium-binding domain to 18 AA C-terminal of the calcium-binding domain, based on annotation by uniprot.org of the second, third, and fifth FG–GAP domains in M. musculus LINKIN.
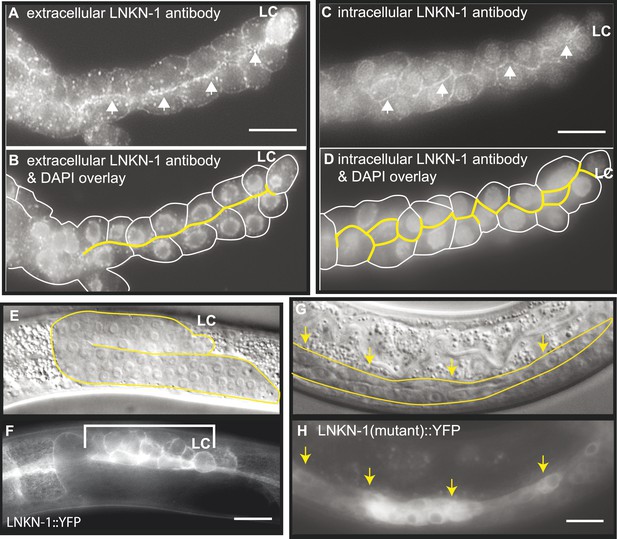
LNKN-1 localizes to the apical and lateral plasma membrane.
(A–D) Immunofluorescence staining of dissected male gonads using antibodies against LNKN-1 extracellular domain (A) and intracellular domain (C) shows localization to the plasma membrane with enrichment at lateral and apical regions (arrows). The antibody against the extracellular domain also labeled cytoplasmic puncta (A), and the antibody against the intracellular domain also labeled the nucleus (C), but these may be due to non-specific staining since they were present in gonads from lnkn-1 RNAi-treated and mutant animals (Figure 3—figure supplement 1). (B) An overlay of the image from (A) and an image of the same gonad stained with DAPI. (D) An overlay of the image from (C) and an image of the same gonad stained with DAPI. In both (B) and (D), the cells are outlined in white and the apical domain is highlighted in yellow. (E and F) Nomarski and epifluorescence images of a live animal expressing YFP-tagged LNKN-1 show that LNKN-1::YFP is expressed in the plasma membrane of gonadal cells but has spread to the basolateral domain. Bracket marks the male somatic gonad. (G and H) LNKN-1(mutant)::YFP, in which wild-type lnkn-1 cDNA construct from (F) is replaced by lnkn-1(gk367) mutant cDNA, does not localize to the plasma membrane. Nomarski (G) and epifluorescence (H) images are of male somatic gonad from a live animal expressing LNKN-1(mutant)::YFP. Scale bar represents 10 μm. Anterior is to the left, posterior is to the right, dorsal is to the top, and ventral is to the bottom.

LNKN-1 antibodies specifically label LNKN-1 protein in the plasma membrane.
Representative epifluorescence images show dissected male gonads from lnkn-1 RNAi-treated (A), lnkn-1 mutant (B), or wild-type (C) males stained with antibody against the extracellular domain of C. elegans LNKN-1. Gonads in (A), (B), and (C) were treated to the same staining conditions and the images were taken with a 200 ms exposure time. Gonads from lnkn-1 RNAi-treated (D), lnkn-1 mutant (F), and wild-type (E and G) males were stained with an antibody against the intracellular domain of C. elegans LNKN-1 under identical conditions. Images of lnkn-1 RNAi-treated (D) and its control wild-type (E) gonads were taken with 100 ms exposure time. Images of lnkn-1 mutant (F) and its control wild-type (G) gonads were taken at 200 ms exposure time. Plasma membrane staining, particularly in the apical domain (arrows), is reduced in RNAi-treated animals and eliminated in lnkn-1 mutants. Scale bar represents 10 μm in all images.

Expression pattern for YFP-tagged LNKN-1.
LNKN-1::YFP localizes to the plasma membrane. In addition to the male somatic gonad (Figure 3E,F), expression was seen in: (A and B) seam cells, (C and D) pharynx (bracket), pharyngeal-intestinal valve (arrow), and excretory cell (arrowhead), (E and F) hermaphrodite somatic gonad (bracket) and vulval precursor cells (arrows), (G and H) the hermaphrodite distal tip cell, (I and J) intestine (bracket), male hook precursor cells (arrowhead), and tail cells (arrow), and (K and L) excretory canal (arrows, longitudinal fluorescent line) and intestine in the background. (M and N) YFP fused to the beginning of the extracellular domain of LNKN-1, in YFP::LNKN-1 constructs, also shows the same plasma membrane localization. Bracket marks the male somatic gonad. (O and P) Dissected, fixed gonad from LNKN-1::YFP expressing animals (O) and the same gonad stained with DLG-1 antibody, an apical domain marker. LNKN-1::YFP is expressed uniformly in the plasma membrane and does not show enrichment in the apical domain as marked by DLG-1 antibody. In contrast, LNKN-1 antibody staining shows enriched LNKN-1 localization to the apical domain (Figure 3A–D). In all figures, the top panel is a Nomarski micrograph and the bottom panel is an epifluorescence image of the same region. Scale bars represent 10 μm except in I and J, where they represent 20 μm. Anterior is left, posterior is right, dorsal is up, and ventral is down.

lnkn-1 RNAi silencing reduces LNKN-1 protein and causes gonad cell detachment defects.
(A) A male treated with lnkn-1 RNAi exhibits gonadal defects in the L4 stage. The gonad (outlined in yellow) stretches thin and detaches from the LC (yellow arrow). (B and C) Nomarski and fluorescence images of a male expressing LNKN-1 tagged with YFP (LNKN-1::YFP) show expression in gonadal cells (outlined in yellow). (D and E) Treatment of LINKIN::YFP expressing animals with lnkn-1 RNAi reduces LINKIN::YFP expression in the gonad (outlined in yellow) to undetectable levels.
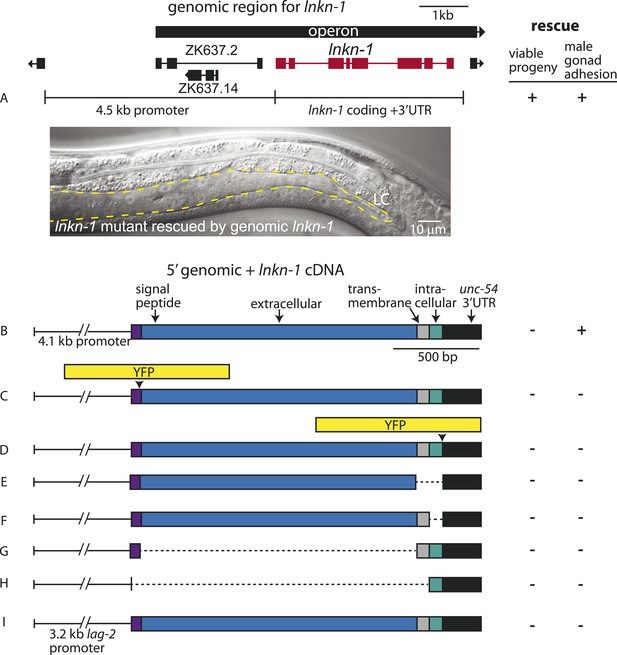
Rescue of lnkn-1 mutant phenotypes requires the full-length lnkn-1 gene.
(A) A full-length genomic construct containing 4.5 kb of 5′ regulatory region, lnkn-1 coding region, and 3′ UTR rescues both gonad detachment defects in the male and maternal lethality. ‘+’ indicates rescue and ‘−’ indicates no rescue. Micrograph shows the posterior body of a male lnkn-1(gk367) mutant that has been rescued for gonad detachment by a full-length genomic lnkn-1 construct. (B) 4.1 kb of genomic promoter region fused to lnkn-1 cDNA and unc-54 3′ UTR rescues male gonad defect but not maternal lethality. (C and D) Constructs with YFP inserted within the extracellular domain (C) or the intracellular domain (D) of lnkn-1 cDNA did not rescue lnkn-1 mutants. (E–H) 4.1 kb of genomic 5′ control region fused to partial domains of lnkn-1(cDNA) do not rescue. lnkn-1 constructs of extracellular domain only (E), extracellular and transmembrane domain (F), transmembrane and intracellular domain (G), and intracellular only (H) also did not rescue lnkn-1 mutants. (I) LC-specific expression of lnkn-1 using lag-2 control sequences also does not rescue.
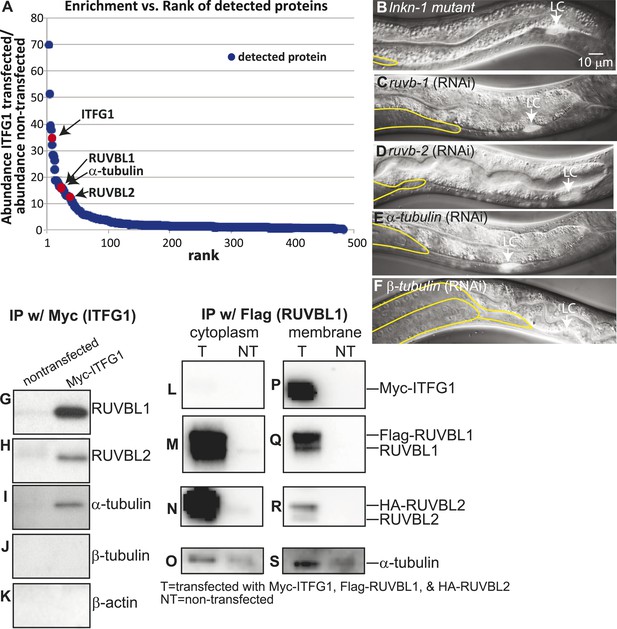
Interactors of LINKIN are RUVBL1, RUVBL2, and α-tubulin.
(A) Graph represents human LINKIN interactors identified by mass spectrometry. ITFG1 (human LINKIN) co-immunoprecipitates from SILAC treated HEK 293T cells with ITFG1-Myc expression were compared to unlabeled cells without ITFG1-Myc expression. (B–F) RNAi knockdowns in C. elegans of ruvb-1, ruvb-2, α-tubulin, and β-tubulin show the same gonad cell detachment as lnkn-1 mutant. Gonad is outlined in yellow and LC is marked by cytoplasmic YFP. Figures are an overlay of Nomarski and fluorescence images. (G–K) Western blots of ITFG1 (human LINKIN) co-immunoprecipitates probed with antibodies against RUVBL1, RUVBL2, α-tubulin, β-tubulin, and control β-actin show that LINKIN interacts with RUVBL1, RUVBL2, and α-tubulin. Myc immunoprecipitation was performed on non-transfected cells (left column) and ITFG1-Myc transfected cells (right column). Equal protein loading was determined by Ponceau S staining. (L–S) Lysates from cells transfected with Myc-ITFG1, Flag-RUVBL1, and HA-RUVBL2 (labeled ‘T’) or with Myc control (labeled ‘C’) were separated into a cytoplasmic and a membrane fraction. The cytoplasmic fraction (L–O) and membrane fraction (P–S) were immunoprecipitated using Flag-RUVBL1 and assayed by Western blot for interactors. ITFG1 is only detected in the membrane fraction (P). RUVBL1 (M), RUVBL2 (N), and α-tubulin (O) interact in the cytoplasmic fraction even without ITFG1. ITFG1 (P), RUVBL1 (Q), RUVBL2 (R), and α -tubulin (S) interact in the membrane fraction.
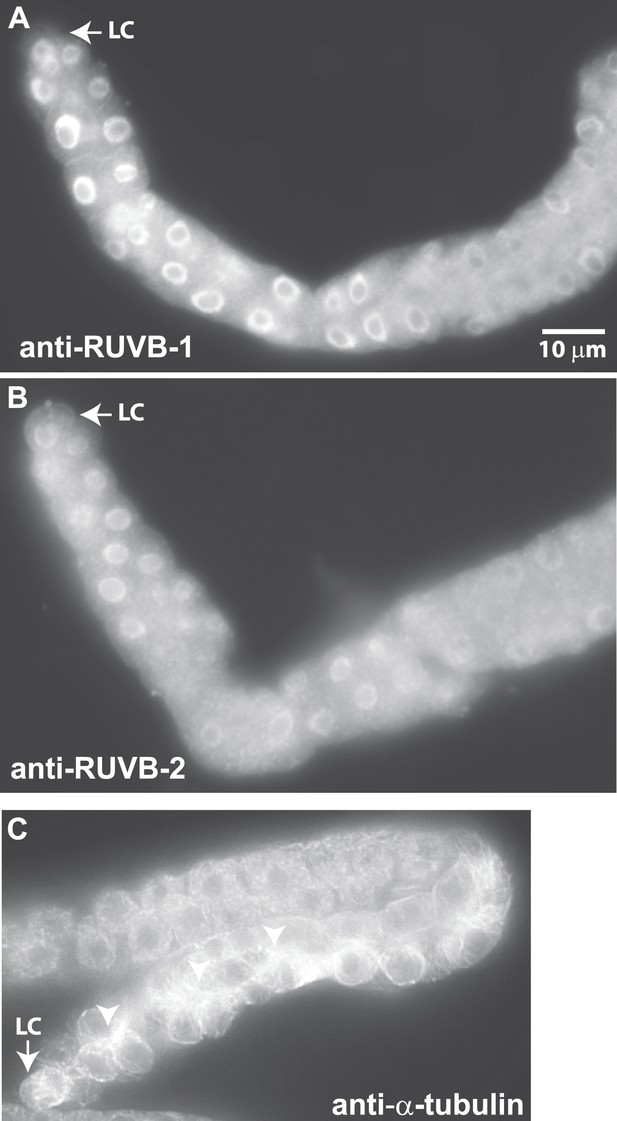
Antibodies against RUVB-1, RUVB-2, and α-tubulin show localization in C. elegans male gonads.
Dissected male gonads stained with antibody against C. elegans RUVB-1 (A) and RUVB-2 (B) show localization in cytoplasm and nucleus. Figure 6—figure supplement 1 shows that both cytoplasmic and nuclear stainings are specific to RUVB-1 or RUVB-2 proteins and can be reduced by ruvb-1 or ruvb-2 RNAi. (C) Dissected male gonad stained with antibody against α-tubulin shows network of microtubule fibers throughout the gonad, with stronger localization to the cell cortex and apical domain (arrow).
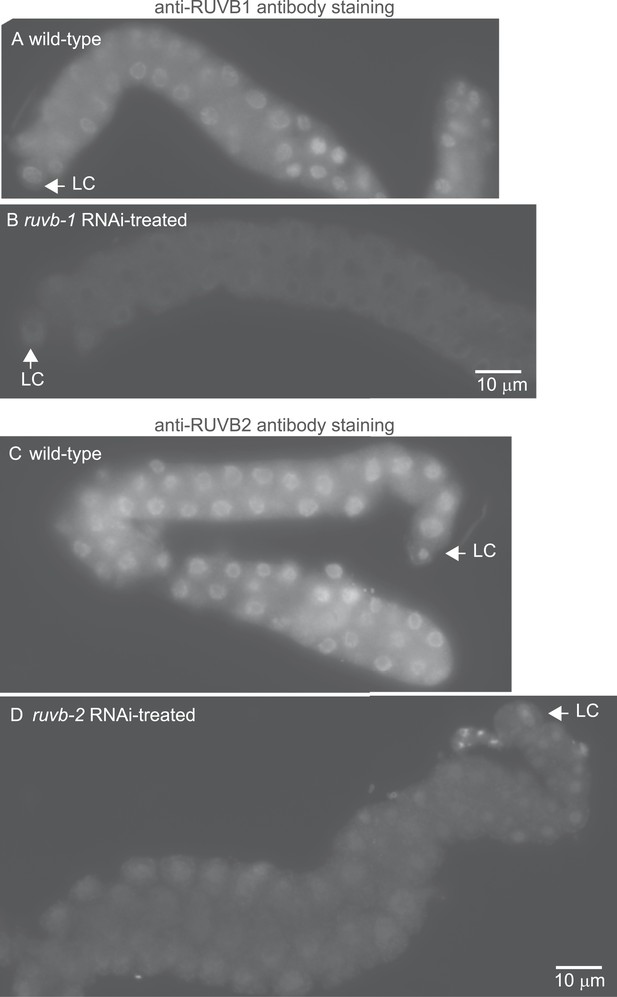
The anti-RUVB-1 antibody specifically labels RUVB-1 protein.
Representative epifluorescence images show dissected gonads from wild-type (A) or ruvb-1 RNAi-treated (B) males stained with antibody against C. elegans RUVB-1 and gonads from wild-type (C) or ruvb-2 RNAi-treated (D) males stained with antibody against C. elegans RUVB-2. The pairs of gonads in A and B and the gonads in C and D were treated to identical staining conditions and imaged at the same exposure times. Both the cytoplasmic and nuclear staining are reduced in RNAi-treated animals.
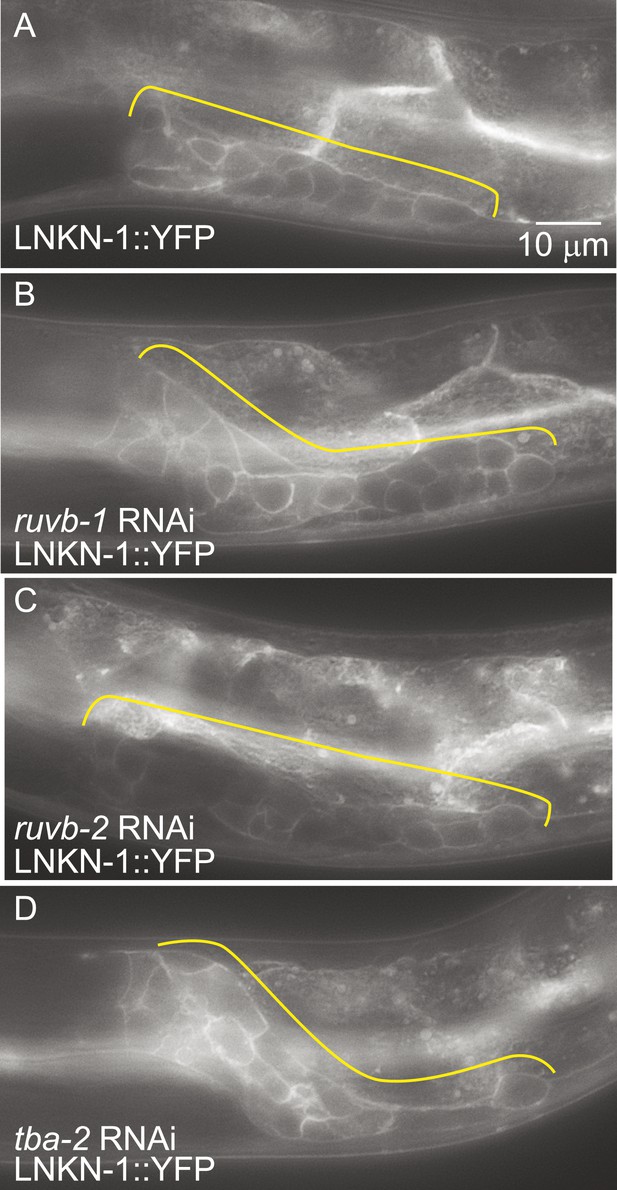
LNKN-1 expression and localization are not dependent on ruvb-1, ruvb-2, or tba-2 function.
Representative epifluorescence images of LINKIN::YFP animals that were either not treated (A), or treated with RNAi against ruvb-1 (B), ruvb-2 (C), or tba-2 (D). The expression of LINKIN::YFP and its localization to the plasma membrane is similar in RNAi-treated and untreated animals.
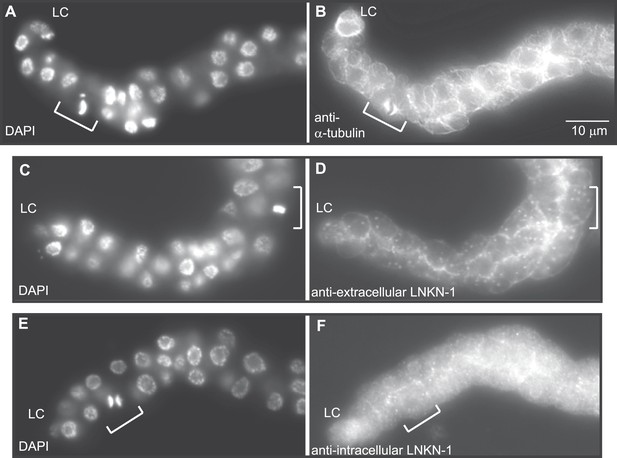
LNKN-1 remains at the plasma membrane during cell division.
(A, C, E) In DAPI-stained dissected male gonads, dividing cells (white bracket) were identified by their condensed chromosomes. (B) Anti-α-tubulin staining shows that microtubules redistribute during cell division to radiate out from the mitotic spindle. (D and F) Staining with anti-LNKN-1 antibody against either the extracellular domain (D) or intracellular domain (F) shows that LNKN-1 remains at the membrane during cell division. LNKN-1 localization is the same in dividing cells (white bracket) and non-dividing neighbors.
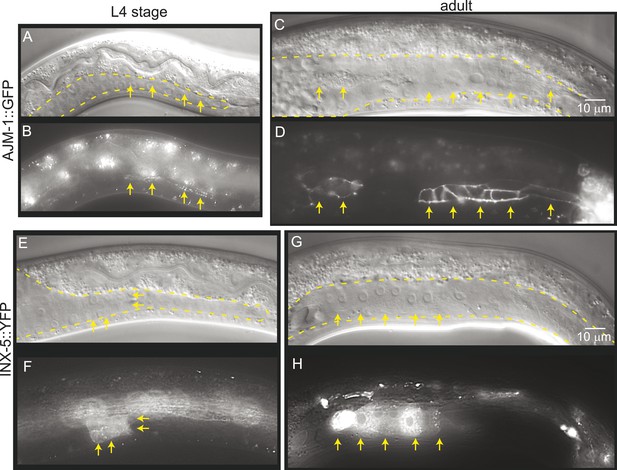
Mature cell–cell junctions form during the L4 stage.
(A and B) Adherens junction marker, AJM-1::GFP, begins to localize as puncta to the apical region of the gonad in the L4 stage (arrows). (C and D) In the adult gonad, AJM-1::GFP lines the apical junctions. (E and F) INX-5::YFP, an innexin expressed in the male gonad, begins to be expressed and accumulate at gap junctions in the L4 stage in a cluster of somatic gonadal cells (arrow). (G and H) INX-5::YFP expression becomes stronger in the adult. Neither AJM-1::GFP nor INX-5::YFP expresses in the gonad in the L3 stage. For each image pair, the top panel is a Nomarski image and bottom is a fluorescence image.
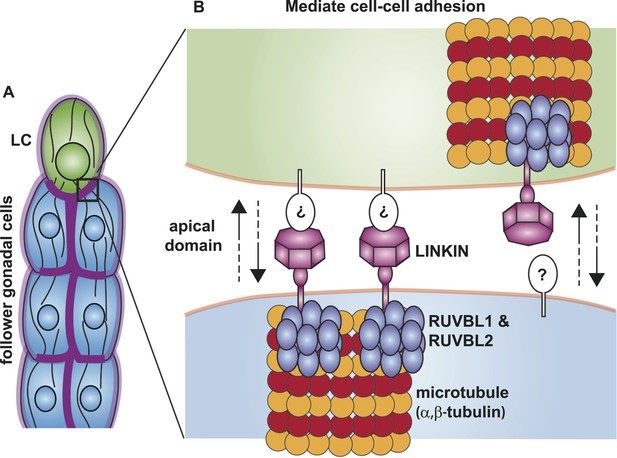
Model for the function of LINKIN, RUVBL1, RUVBL2, and microtubule proteins in cell–cell adhesion.
(A) The male gonad shape is generated by a collective migration of the leader LC (green) and follower somatic cells (blue). LINKIN (purple) is a transmembrane glycoprotein expressed in the plasma membrane of all gonadal cells, with enrichment at apical and lateral domains (dark purple). LNKN-1 is required for cell–cell adhesion during gonadal migration. (B) The interface between two adherent cells boxed in (A) is shown in more detail. LINKIN integrates interactions with neighboring cells on the cell exterior with connections to the microtubule cytoskeleton on the cell interior. On the extracellular side, the β-propeller domain (purple heptagon) of LINKIN binds an unidentified partner (white oval) on the adjacent cell membrane. The highly conserved intracellular domain of LINKIN binds RUVBL1, RUVBL2, and α-tubulin at the intracellular face of the plasma membrane. Based on RUVBL1 and RUVBL2's ability to form stacked hetero-hexameric rings (Gorynia et al., 2011) and regulate microtubule nucleation and dynamics (Gartner et al., 2003), we propose that they assist in interaction between LINKIN and microtubules.
Additional files
-
Supplementary file 1
Interactors of human LINKIN identified by mass spectrometry.
- https://doi.org/10.7554/eLife.04449.017
-
Supplementary file 2
Top interactors of human LINKIN identified by mass spectrometry (>fivefold enriched over control).
- https://doi.org/10.7554/eLife.04449.018
-
Supplementary file 3
lnkn-1 constructs used for rescuing lnkn-1 mutant.
- https://doi.org/10.7554/eLife.04449.019





