Centriolar satellites assemble centrosomal microcephaly proteins to recruit CDK2 and promote centriole duplication
Figures
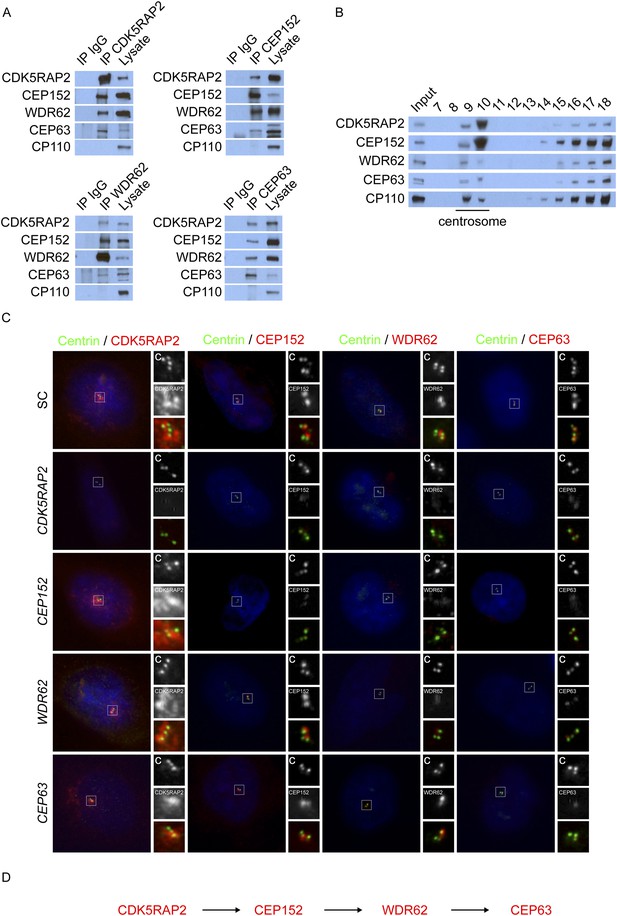
CDK5RAP2, CEP152, WDR62, and CEP63 interact at the centrosome and localize to centrioles in a stepwise manner.
(A) We immunoprecipitated endogenous CDK5RAP2, CEP152, WDR62, and CEP63 from HeLa total cell lysates. Precipitation and co-precipitation were detected using antibodies specific to CDK5RAP2, CEP152, WDR62, and CEP63. The centriole component CP110 served as a negative control. (B) Sucrose gradient fractions of HeLa cell lysates were analyzed by immunoblot with antibodies to CP110, CDK5RAP2, CEP152, WDR62, and CEP63. (C) Immunofluorescence of S phase scrambled control (SC), CDK5RAP2, CEP152, WDR62, and CEP63 siRNA-treated HeLa cells co-stained for Centrin (‘c’, green) to visualize centrioles, CDK5RAP2 (red), CEP152 (red), WDR62 (red), and CEP63 (red), and nuclei (DAPI, blue). The inset shows magnified images of the boxed region. (D) Our findings indicate that CDK5RAP2, recruits CEP152 to the centrosome, which in turn recruits WDR62 and CEP63. Scale bars indicate 5 μm for all images.
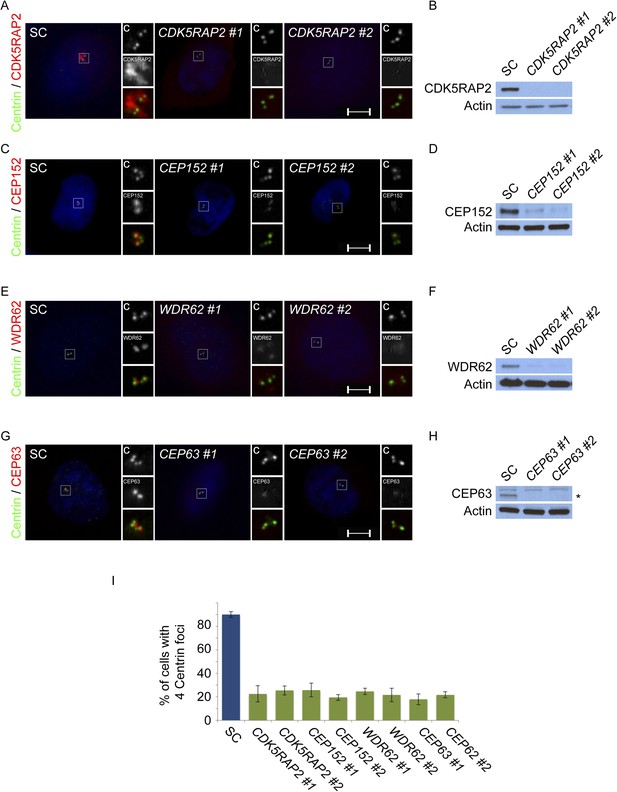
CDK5RAP2, CEP152, WDR62 and CEP63 are required for centriole duplication.
(A) SC, CDK5RAP2 #1, or CDK5RAP2 #2 siRNA-treated S phase HeLa cells were analyzed by immunofluorescence with Centrin (‘c’, green) and CDK5RAP2 (red). (B) Immunoblotting of SC, CDK5RAP2 #1, or CDK5RAP2 #2 siRNA transfected HeLa cell total cell lysate analyzed with an antibody to CDK5RAP2. (C) Immunofluorescence images of S phase HeLa cells transfected with SC, CEP152 #1, or CEP152 #2 siRNA and co-stained with Centrin (‘c’, green) and CEP152 (red). (D) Total cell lysate of SC, CEP152 #1, or CEP152 #2 siRNA-treated HeLa cells was analyzed by immunoblot with an antibody to CEP152. (E) S phase HeLa cells transfected with SC, WDR62 #1, or WDR62 #2 siRNA were co-stained with Centrin (‘c’, green) and WDR62 (red). (F) Unboiled total cell lysate of SC, WDR62 #1, or WDR62 #2 siRNA-treated HeLa cells was analyzed by immunoblot with an antibody to WDR62. (G) Immunofluorescence analysis of SC, CEP63 #1, or CEP63 #2 siRNA transfected HeLa cells co-stained with Centrin (‘c’, green) and CEP63 (red). (H) Immunoblot of total cell lysate of HeLa cells transfected with SC, CEP63 #1, or CEP63 #2 siRNA and analyzed with an antibody to CEP63. (I) Quantification of S phase SC, CDK5RAP2 #1, CDK5RAP2 #2, CEP152 #1, CEP152 #2, WDR62 #1, WDR62 #2, CEP63 #1, or CEP63 #2 siRNA-treated HeLa cells with four centrioles. S phase cells were identified by CyclinA immunostaining. For all quantifications at least 100 cells were counted per experiment (n = 3), p < 0.005 (paired t-test). Actin served as a loading control for all immunoblot analyses. Scale bars indicate 5 μm for all images.
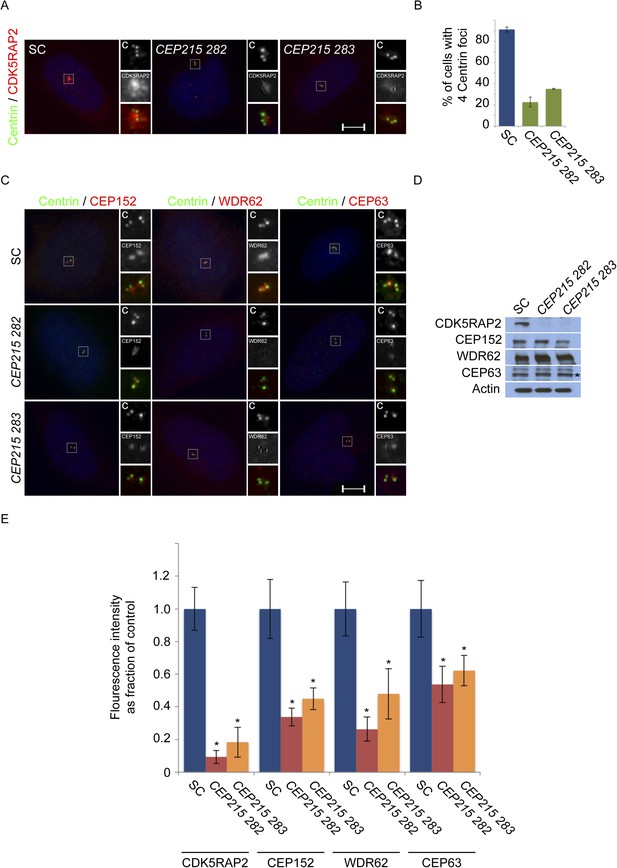
CDK5RAP2/CEP215 promotes centriole duplication and centrosome organization.
(A) S phase SC, CEP215 282, CEP215 283 siRNA-treated HeLa cells co-stained with Centrin (‘c’, green) and CDK5RAP2 (red). (B) Quantification of S phase SC, CEP215 282, CEP215 283 siRNA transfected HeLa cells with four centrioles. For all quantifications at least 100 cells were counted per experiment (n = 3), p < 0.005 (paired t-test). (C) Immunofluorescence images of S phase HeLa cells transfected with SC, CEP215 282, CEP215 283 siRNA and co-stained with Centrin (‘c’, green) and CEP152 (red), WDR62 (red), or CEP63 (red). Scale bars indicate 5 μm for all images. (D) Immunoblot of total cell lysate of HeLa cells transfected with SC, CEP215 282, CEP215 283 siRNA and analyzed with antibodies to CDK5RAP2, CEP152, WDR62, and CEP63. Actin served as a loading control. (E) Quantification of the mean fluorescence intensities ±s.d. of CDK5RAP2, CEP152, WDR62 and CEP63 in SC, CEP215 282, CEP215 283 siRNA-treated cells expressed as the mean percentage ±s.d. of the fluorescence intensities of SC cells. For fluorescence quantifications 30 cells were analyzed per experiment (n = 3). p < 0.005 (paired t-test) statistically significant differences are denoted by an asterisk.
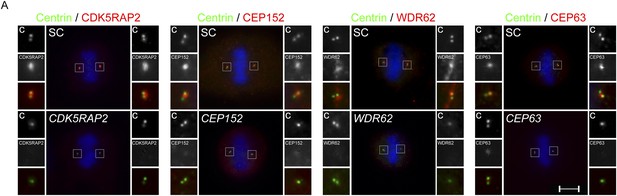
Centriole duplication defects in CDK5RAP2, CEP152, WDR62 and CEP63-depleted mitotic cells.
(A) SC, CDK5RAP2, CEP152, WDR62 and CEP63 siRNA treated mitotic HeLa cells were co-stained for Centrin (‘c’, green) and CDK5RAP2 (red), CEP152 (red), WDR62 (red), or CEP63 (red). Scale bars indicate 5 μm for all images.
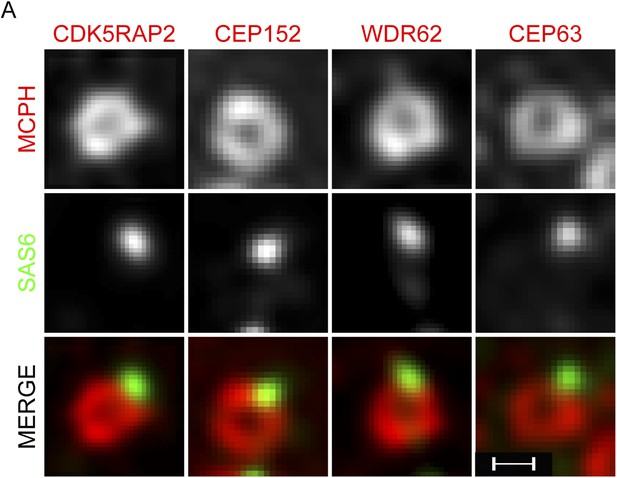
CDK5RAP2, CEP152, WDR62 and CEP63 form a ring near parental centrioles.
(A) HeLa cells were co-stained with antibodies to SAS6 (green), CDK5RAP2 (red), CEP152 (red), WDR62 (red), and CEP63 (red) and imaged using three-dimensional structured illumination microscopy (SIM). Scale bars indicate 200 nm for all images.
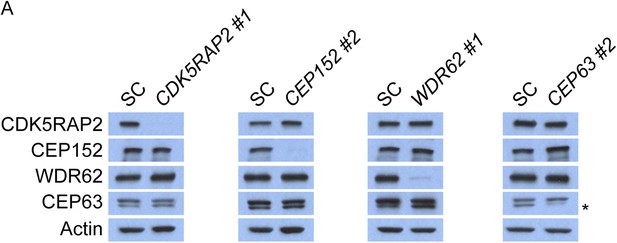
CDK5RAP2, CEP152, WDR62 and CEP63 do not control the stability of their binding partners.
(A) Total cell lysate of SC, CDK5RAP2, CEP152, WDR62 and CEP63 siRNA transfected HeLa cells were analyzed by immunoblotting with antibodies to CDK5RAP2, CEP152, WDR62 and CEP63. Actin served as a loading control.
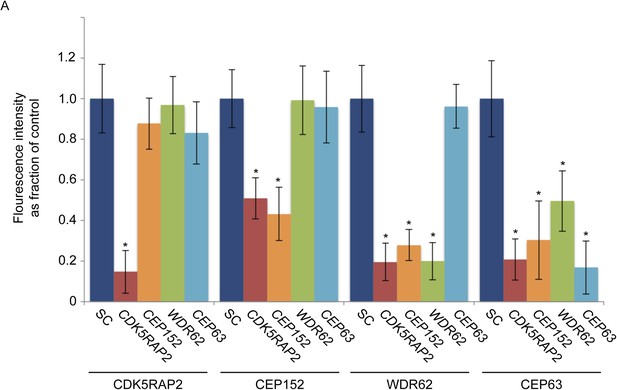
CDK5RAP2, CEP152, WDR62 and CEP63 localize in a hierarchical manner at the centrosome.
(A) Quantification of CDK5RAP2, CEP152, WDR62 and CEP63 in SC, CDK5RAP2, CEP152, WDR62, CEP63-depleted cells expressed as the mean percentage ±s.d. of the fluorescence intensities of SC cells. For all quantifications 30 cells were analyzed per experiment (n = 3). p < 0.005 (paired t-test) statistically significant differences from the SC control cells are denoted by an asterisk.
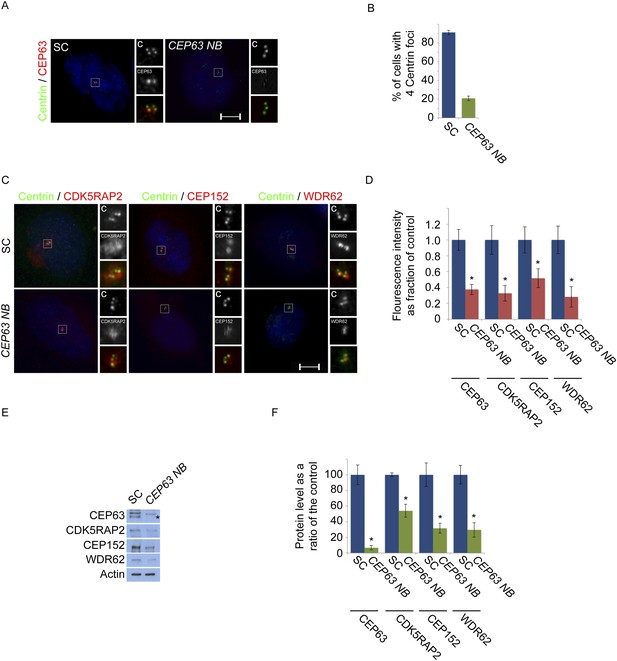
CEP63-depletion using a previous published siRNA destabilizes MCPH proteins.
(A) HeLa cells in S phase transfected with SC or CEP63 Nicola Brown (NB) siRNA were co-stained with Centrin (‘c’, green) and CEP63 (red). (B) Quantification of the mean percentage of SC and CEP63 NB siRNA treated cells in S phase with four centrioles. (C) S phase HeLa cells transfected with SC and CEP63 NB siRNA were co-stained with Centrin (‘c’, green) and CDK5RAP2 (red), CEP152 (red), or CEP63 (red). (D) Quantification of the mean fluorescence intensities ±s.d. of CDK5RAP2, CEP152, WDR62 and CEP63 in SC and CEP63 NB siRNA treated cells expressed as the mean percentage ±s.d. of the fluorescence intensities of SC cells. For all quantifications 30 cells were analyzed per experiment (n = 3). p < 0.005 (paired t-test) statistically significant differences from SC controls are denoted by an asterisk. (E) Total cell lysates of SC and CEP63 NB siRNA treated cellswere analyzed by Western blot with antibodies to CEP63, CDK5RAP2, CEP152, and WDR62. Actin served as a loading control. Asterisk indicates the specific band. (F) Quantification of the mean intensities ±s.d. of CEP63, CDK5RAP2, CEP152, and WDR62 in SC and CEP63 NB siRNA-treated cells expressed as the mean percentage ±s.d. of the intensities of SC samples. Actin served as a loading control and to normalize samples (n = 3). Asterisk denotes statistical significance compared to controls where p < 0.005 (paired t-test).
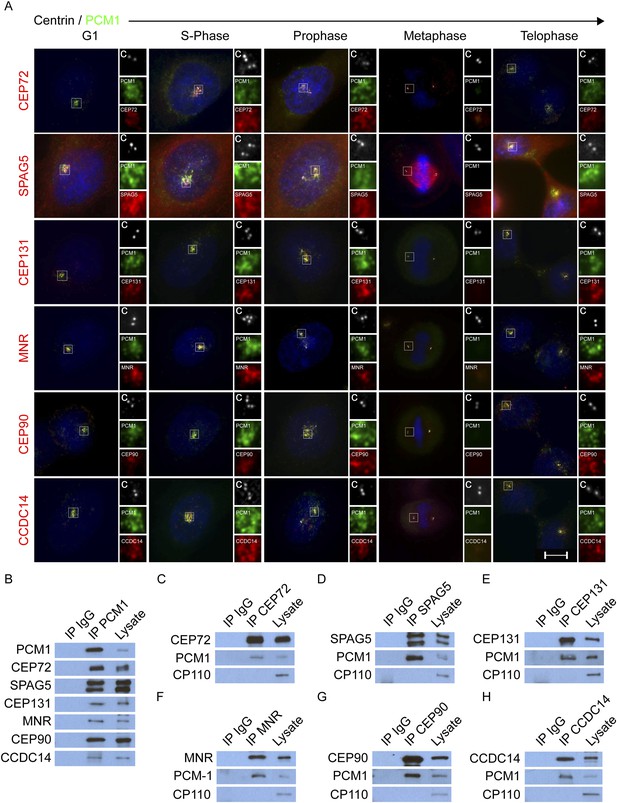
CEP72, SPAG5, CEP131, MNR, CEP90 and CCDC14 are centriolar satellite components.
(A) Immunofluorescence microscopy of asynchronously growing HeLa cells co-stained with antibodies to Centrin (‘c’, white), PCM1 (green), CEP72 (red), SPAG5 (red), CEP131 (red), MNR (red), CEP90 (red), CCDC14 (red). Cell cycle stage was indicated by the number of Centrin (‘c’, green) foci and DNA condensation (Hoechst, blue). The insets show magnified images of the boxed regions. Scale bars indicate 5 μm for all images. (B) HeLa total cell lysates were subjected to immunoprecipitation of PCM1 and c-Myc (IgG), which served as a negative control throughout. Precipitating proteins were subjected to immunoblotting for PCM1, CEP72, SPAG5, CEP131, MNR, CEP90, and CCDC14. (C–H) Reciprocal immunoprecpitation of PCM1 and copurified proteins. Endogenous proteins were immunoprecipitated with antibodies to CEP72, SPAG5, CEP131, MNR, CEP90, CCDC14, and a negative control IgG. Complexes were immunoblotted with antibodies to PCM1, CEP72, SPAG5, CEP131, MNR, CEP90, and CCDC14. Scale bars indicate 5 μm for all images.
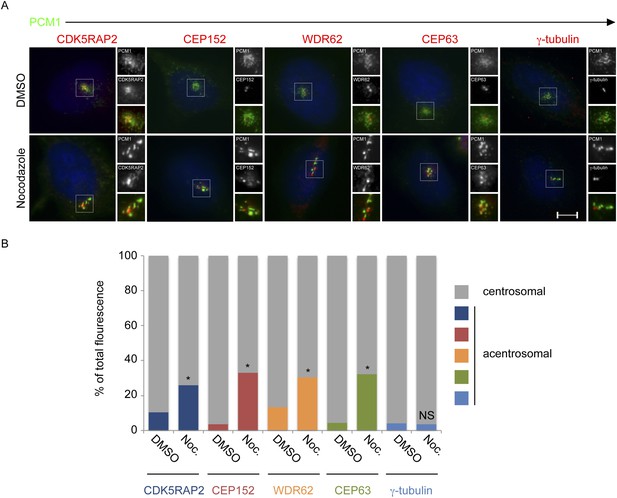
CDK5RAP2, CEP152, WDR62 and CEP63, but not γ-tubulin, localize to the centrosome in a microtubule-dependent manner.
(A) HeLa cells treated with DMSO or 17 μM nocodazole were analyzed by immunofluorescence for PCM1 (‘p’, green), CDK5RAP2 (red), CEP152 (red), WDR62 (red), CEP63 (red) and γ-tubulin (red). Scale bars indicate 5 μm for all images. (B) Quantification of the relative mean fluorescence intensities of CDK5RAP2, CEP152, WDR62, CEP63 and γ-tubulin at the centrosome (gray) and acentrosomal (as defined by colocalization with the centriolar satellite component PCM1; blue, red, orange, green and teal) in DMSO or nocodazole-treated cells. For all quantifications, 15 cells were analyzed per experiment (n = 3). p < 0.005 (paired t-test) statistically significant differences from DMSO-treated controls are denoted by an asterisk.
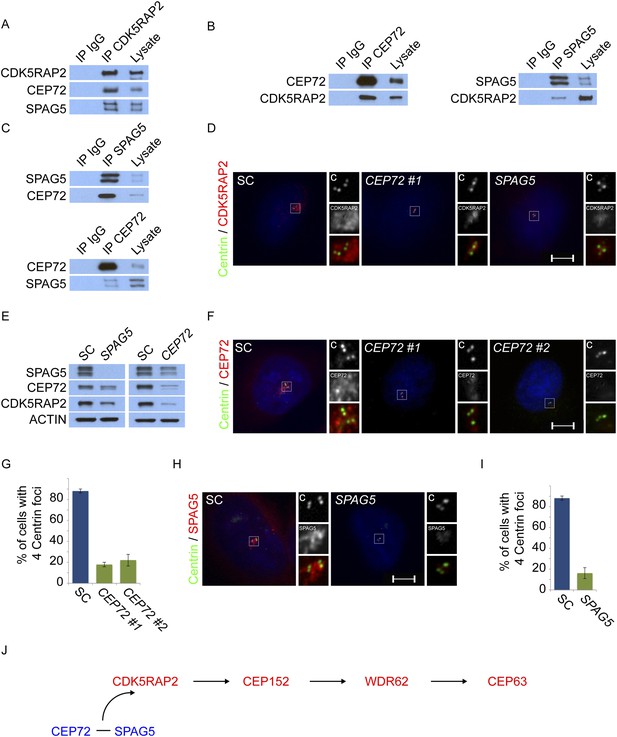
Centriolar satellite components CEP72 and SPAG5 stabilize and localize CDK5RAP2 to the centrosome to promote centriole duplication.
(A) HeLa total cell lysates were subjected to immunoprecipitation of CEP72, SPAG5, and a negative control. Precipitating proteins were subjected to immunoblotting for CEP72, SPAG5 and CDK5RAP2. (B) The interactions between CEP72 and SPAG5 with CDK5RAP2 were validated by immunoprecipitation using an antibody to CDK5RAP2. Endogenous CDK5RAP2 was immunoprecipitated and complexes were immunoblotted with antibodies to CEP72, SPAG5 and CDK5RAP2. (C) Immunoprecipitated CEP72, SPAG5, and a negative control were immunoblotted using antibodies to CEP72, SPAG5 and a negative control. (D) HeLa cells in S phase transfected with SC, CEP72 #1 or SPAG5 siRNA were co-stained for CDK5RAP2 (red) and Centrin (‘c’, green) to visualize centrioles. (E) Total cell lysates of SC, CEP72 #1, and SPAG5 siRNA transfected HeLa cells were analyzed by immunoblotting with antibodies to CEP72, SPAG5, and CDK5RAP2. Actin served as a loading control. (F) SC, CEP72 #1, CEP72 #2 siRNA transfected HeLa cells were co-stained with CEP72 (red) and Centrin (‘c’, green) to visualize centrioles. (G) Percentage of S phase cells with four Centrin foci in SC, CEP72 #1, CEP72 #2 siRNA treated HeLa cells. Error bars represent ±s.d. throughout. (H) S phase HeLa cells transfected with SC or SPAG5 siRNA co-stained with Centrin (‘c’, green) and SPAG5 (red). (I) Quantification of S phase SC and SPAG5-depleted HeLa cells with four Centrin foci. (J) Our findings suggest that CEP72 and SPAG5 are required to localize CDK5RAP2 to the centrosome, which itself recruits CEP152, WDR62 and CEP63. Centrosome localized MCPH proteins are represented in red and centriolar satellites in blue. For all quantifications at least 100 cells were counted per experiment (n = 3), p < 0.005 (paired t-test) for SC vs CEP72 #1, #2 and SPAG5 siRNA transfected cells. Scale bars indicate 5 μm for all images.
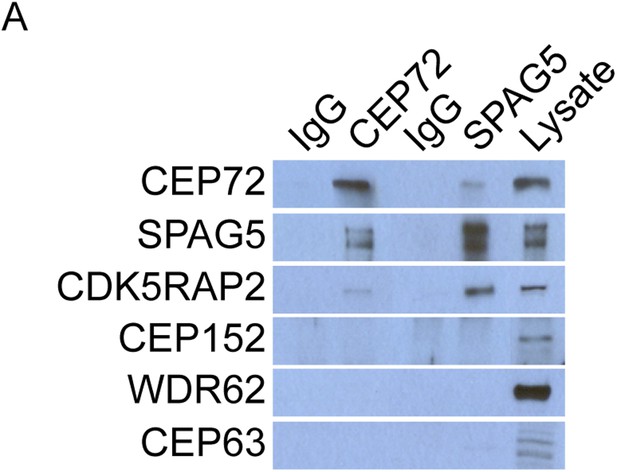
CEP72 and SPAG5 interact with CDK5RAP2 but not CEP152, WDR62 or CEP63.
(A) HeLa total cell lysates were immunoprecipitated with antibodies to CEP72 and SPAG5. Co-precipitating proteins were probed with antibodies to CEP72, SPAG5, CDK5RAP2, CEP152, WDR62, and CEP63.
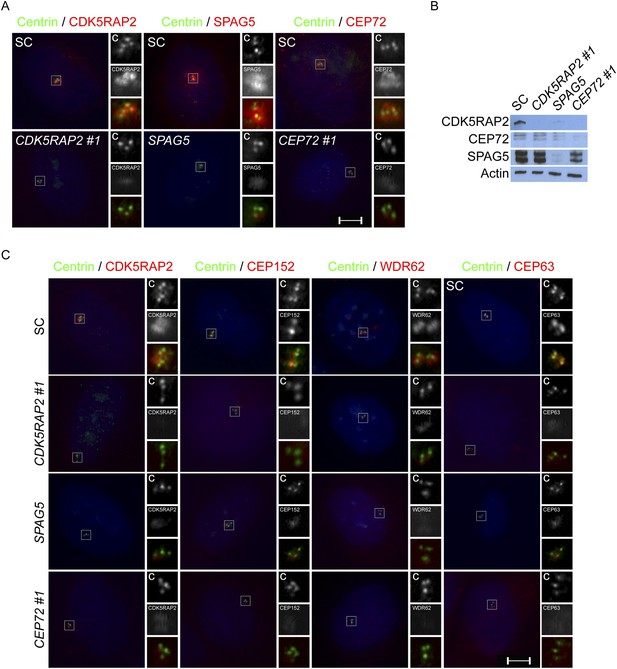
CEP72, SPAG5 and CDK5RAP2 promote centriole duplication and centrosome organization in U2OS cells.
(A) SC, CDK5RAP2, CEP72 and SPAG5-depleted S phase U2OS cells were co-stained with Centrin (‘c’, green), and CDK5RAP2 (red), SPAG5 (red) or CEP72 (red). (B) Total cell lysate from SC, CEP72 and SPAG5 siRNA-treated U2OS cells were analyzed by western blotting using antibodies to CDK5RAP2, CEP72, and SPAG5. Actin served as a loading control. (C) S phase SC, CDK5RAP2, CEP72, and SPAG5 siRNA-treated U2OS cells were co-stained with Centrin (‘c’, green), CDK5RAP2 (red), CEP152 (red), WDR62 (red), and CEP63 (red). Scale bars indicate 5 μm for all images.
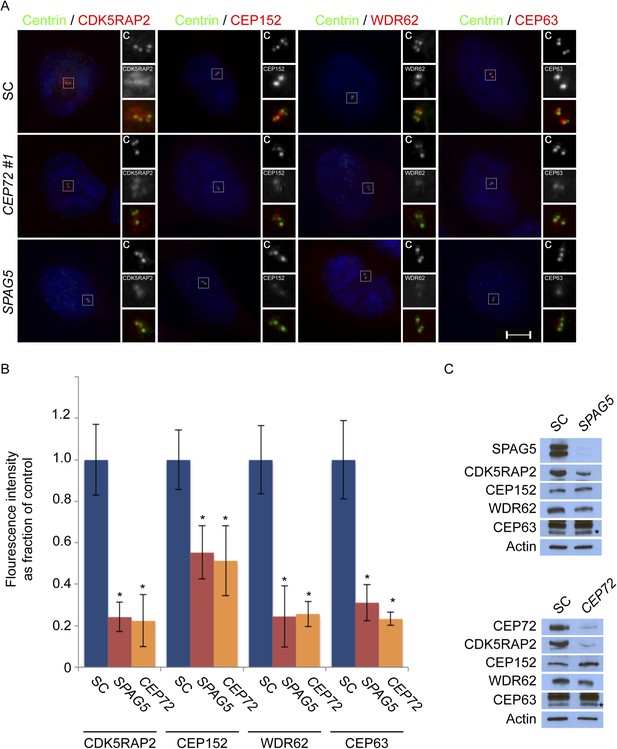
CEP72 and SPAG5 are required to localize CDK5RAP2, CEP152, WDR62 and CEP63 to the centrosome.
(A) S phase SC, CEP72 #1, and SPAG5 siRNA-treated HeLa cells were co-stained with Centrin (‘c’, green), CDK5RAP2 (red), CEP152 (red), WDR62 (red), and CEP63 (red). Scale bar indicates 5 μm. (B) Quantification of the intensities of centrosomal CDK5RAP2, CEP152, WDR62 and CEP63 in SC, SPAG5 and CEP72 siRNA-treated cells expressed as the mean percentage ±s.d. of the intensities of SC samples. For fluorescence quantifications 30 cells were analyzed per experiment (n = 3). p < 0.005 (paired t-test) statistically significant differences are denoted by an asterisk. (C) Total cell lysates of SC SPAG5 and CEP72 siRNA treated cells were analyzed by immunoblot with antibodies to CDK5RAP2, CEP152, WDR62 and CEP63. Actin served as a loading control. Asterisk indicates the specific band.
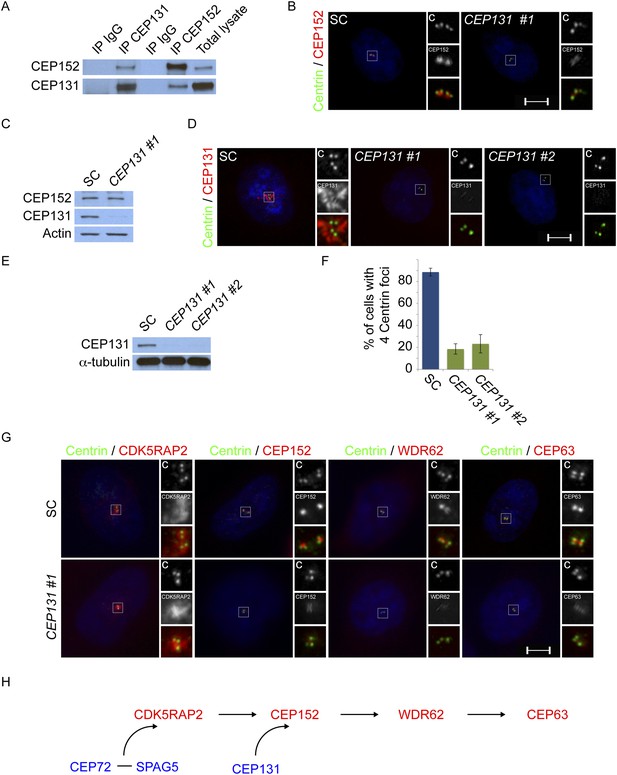
Centriolar satellite component CEP131 interacts with and localizes CEP152 to the centrosome to promote centriole duplication.
(A) HeLa total cell lysates were subjected to immunoprecipitation of CEP131, CEP152 and a negative control. Precipitating proteins were subjected to immunoblotting for CEP131 and CEP152. (B) HeLa cells in S phase transfected with SC or CEP131 #1 siRNA were co-stained for CEP152 (red) and Centrin (‘c’, green). (C) Total cell lysates of SC and CEP131 #1 siRNA transfected HeLa cells were analyzed by immunoblotting with antibodies to CEP131 and CEP152. Actin served as a loading control. (D) HeLa cells in S phase transfected with SC, CEP131 #1, or CEP131 #2 siRNA were co-stained for CEP131 (red) and Centrin (‘c’, green) to visualize centrioles. (E) Total cell lysates of SC, CEP131 #1, CEP131 #2 siRNA transfected HeLa cells were analyzed by immunoblotting for Cep131. α-tubulin served as a loading control. 20 μg of protein lysate was loaded per lane. (F) Quantification of the mean percentage of SC, CEP131 #1, or CEP131 #2 siRNA transfected cells in S phase with four centrioles. (G) SC and CEP131-depleted S phase cells were co-stained with Centrin (‘c’, green), CDK5RAP2 (red), CEP152 (red), WDR62 (red), and CEP63 (red). (H) Schematic indicating that CEP131 is required to localize CEP152, WDR62 and CEP63 to the centrosome. For all quantifications at least 100 cells were counted per experiment (n = 3), p < 0.005 (paired t-test). Scale bars indicate 5 μm for all images.
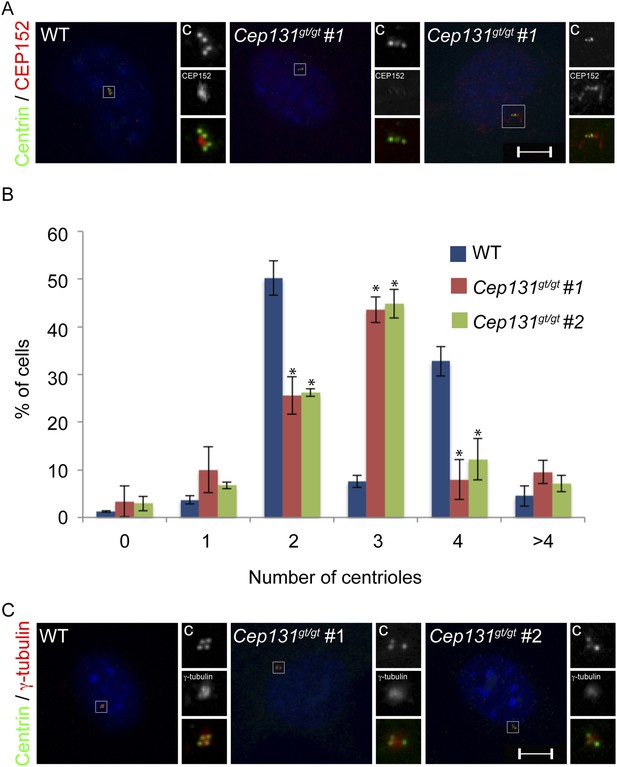
Cep131gt/gt MEFs exhibit centriole duplication and centrosome organizational defects.
(A) WT and two Cep131gt/gt MEF cell lines were analyzed by immunofluorescence with antibodies to Centrin (‘c’, green) and γ-tubulin (red). (B) Quantification of the number of centrioles in asynchronous populations of WT and two Cep131gt/gt MEFs. (C) Immunofluorescence of WT and Cep131gt/gt MEFs co-stained with Centrin (‘c’, green) and CEP152 (red). For all quantifications at least 100 cells were counted per experiment (n = 3). p < 0.005 (paired t-test) compared to WT was denoted with an asterisk. Scale bars indicate 5 μm for all images.
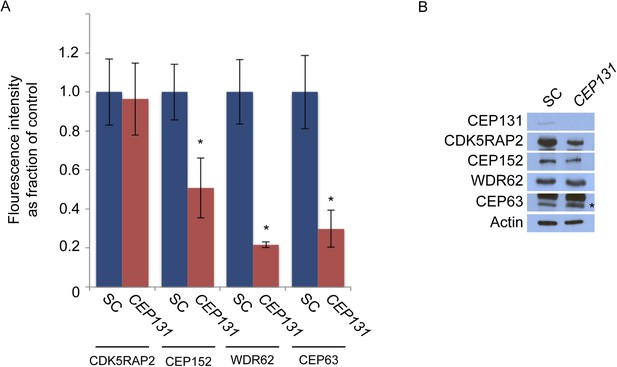
CEP131 is required for the centrosomal localization of CEP152, WDR62 and CEP63.
(A) Quantification of the mean fluorescence intensities ±s.d. of CDK5RAP2, CEP152, WDR62 and CEP63 in SC or CEP131-depleted cells expressed as the mean percentage ±s.d. of the fluorescence intensities of SC cells. For all quantifications 30 cells were analyzed per experiment (n = 3). p < 0.005 (paired t-test) statistically significant differences are denoted by an asterisk. (B) Total cell lysate from SC and CEP131 siRNA-treated HeLa cells were analyzed by immunoblotting using antibodies to CEP131, CDK5RAP2, CEP152, WDR62, and CEP63. Actin served as a loading control.
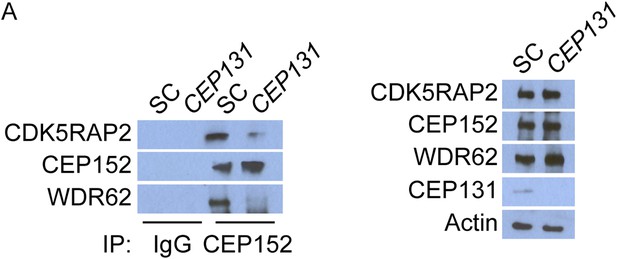
CEP152 associates with CDK5RAP2 and WDR62 in a CEP131-dependent manner.
(A) Total cell lysate from SC and CEP131-depleted HeLa cells were subjected to immunoprecipitation using an antibody to CEP152. Co-precipitating proteins were analyzed by immunoblotting for CDK5RAP2 and WDR62. SC and CEP131 siRNA transfected HeLa cell total cell lysates were analyzed by immunoblotting with antibodies to CDK5RAP2, CEP152, WDR62 and CEP131. Actin served as a loading control.
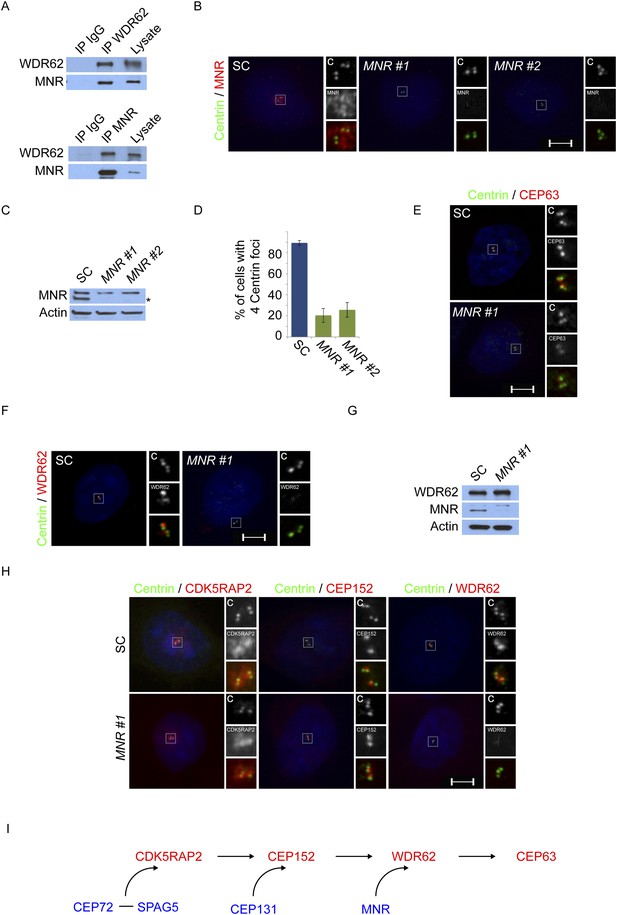
Satellite component MNR promotes centriole duplication by localizing WDR62 to the centrosome.
(A) We immunoprecipitated endogenous WDR62 and MNR from HeLa total cell lysates. Co-precipitation was detected using antibodies specific to WDR62 and MNR. (B) SC, MNR #1, and MNR #2 siRNA transfected S-phase HeLa cells were co-stained with MNR (red) and Centrin (‘c’, green). (C) Total cell lysate of SC, MNR #1, MNR #2 siRNA transfected HeLa cells were analyzed by immunoblotting with antibodies to MNR. The asterisk marks the band specific to MNR, which sits below a non-specific band. Actin served as a loading control. (D) Percentage of S-phase SC, MNR #1, MNR #2 siRNA treated HeLa cells with four Centrin foci. (E) S-phase SC and MNR siRNA-transfected HeLa cells were co-stained for Centrin (‘c’, green) and CEP63 (red). (F) SC and MNR siRNA treated S-phase HeLa cells were co-stained for WDR62 (red) and Centrin (‘c’, green). (G) Total cell lysates of SC and MNR-depleted cells were analyzed by immunoblot with antibodies to WDR62 and MNR. Actin served as a loading control. 20 μg of protein lysate was loaded per lane. (H) SC and MNR #1-depleted S-phase cells were co-stained with Centrin (‘c’, green), CDK5RAP2 (red), CEP152 (red), and WDR62 (red). (I) Our findings indicate that MNR localizes WDR62 to the centrosome, which in turn recruits CEP63. For all quantifications at least 100 cells were counted per experiment (n = 3), p < 0.005 (paired t-test). Scale bars indicate 5 μm for all images.
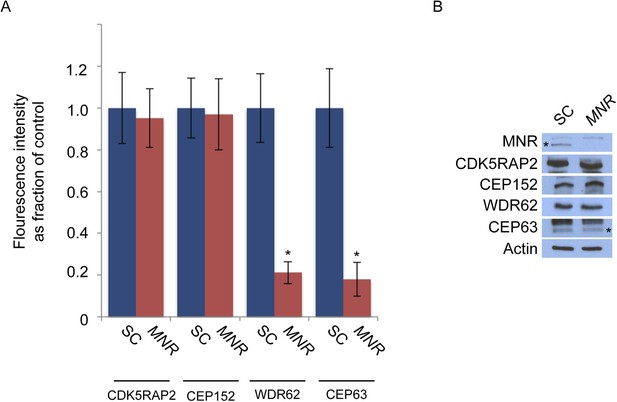
MNR is required to localize WDR62 and CEP63 to the centrosome.
(A) Quantification of the mean fluorescence intensities of centrosomal CDK5RAP2, CEP152, WDR62 and CEP63 in SC or MNR-depleted cells expressed as the mean percentage ±s.d. of the fluorescence intensities of the centrosomal signal of SC cells. For all quantifications 30 cells were analyzed per experiment (n = 3). p < 0.005 (paired t-test) statistically significant differences are denoted by an asterisk. (B) SC and MNR siRNA transfected HeLa cell total cell lysates were analyzed by immunoblotting with antibodies to MNR, CDK5RAP2, CEP152, WDR62 and CEP63. Actin served as a loading control.
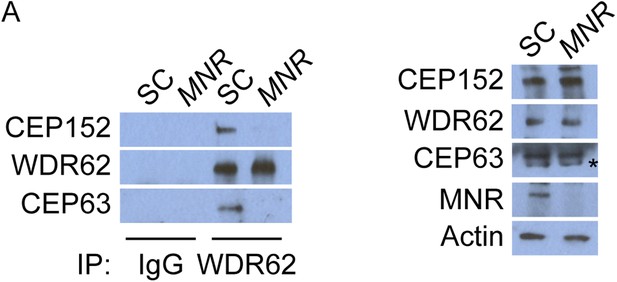
WDR62 interacts with CEP152 and CEP63 in a MNR-dependent manner.
(A) We immunoprecipitated endogenous CEP63 from SC and MNR-depleted HeLa total cell lysates. Precipitation and co-precipitation were detected using antibodies specific to CEP152, WDR62, and CEP63. Total cell lysates from SC and MNR depleted cells were analyzed by immunoblotting with antibodies to CEP152, WDR62, CEP63, and MNR. Actin served as a loading control.
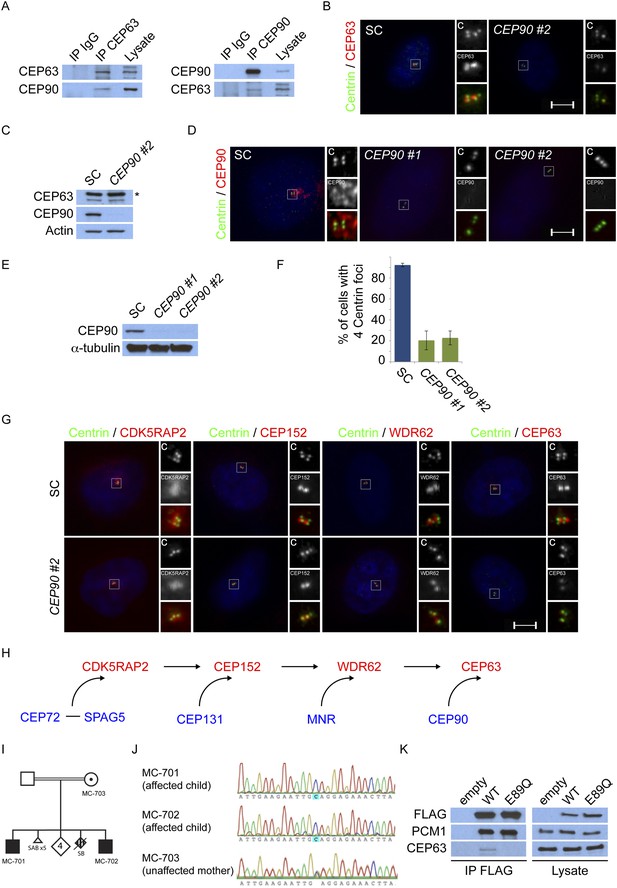
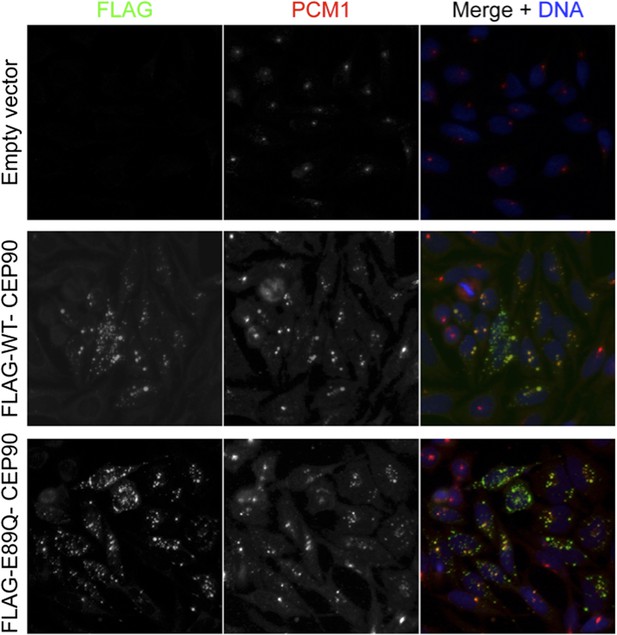
CEP90 encodes a centriolar satellite and MCPH-associated protein required to localize CEP63 to the centrosome to promote centriole duplication.
(A) HeLa total cell lysate was subjected to endogenous immunoprecipitation using antibodies to CEP63, CEP90 and a negative control. Precipitating proteins were analyzed by immunoblotting for CEP63 and CEP90. (B) Immunofluorescence images of SC and CEP90 #2 siRNA-treated HeLa cells co-stained for CEP63 (red) and Centrin (‘c’, green). (C) Total cell lysates of SC or CEP90 #2 siRNA transfected HeLa cells were analyzed by immunoblotting with antibodies to CEP63, CEP90 and Actin, which served as a loading control. Asterisk indicates the specific band. (D) S phase SC, CEP90 #1, and CEP90 #2 siRNA-treated cells were analyzed by immunofluorescence with antibodies to CEP90 (red) and Centrin (‘c’, green). (E) Total cell lysates of SC, CEP90 #1, and CEP90 #2 siRNA transfected HeLa cells were analyzed by immunoblotting for CEP90. α-tubulin served as a loading control. (F) Mean percentage of S phase SC, CEP90 #1, and CEP90 #2 siRNA-treated HeLa cells with four Centrin foci. At least 100 cells were counted per experiment (n = 3), p < 0.005 (paired t-test). (G) SC and CEP90 #2-depleted S phase cells were co-stained with Centrin (‘c’, green), CDK5RAP2 (red), CEP152 (red), WDR62 (red), and CEP63 (red). Scale bars indicate 5 μm for all images. (H) Schematic indicating that CEP90 is required for the centrosomal localization of CEP63. (I) Diagram of a simplified pedigree. Filled squares indicate individuals with MCPH. Spontaneous abortions (SAB), stillbirths (SB), and individuals of unknown gender (diamonds) are also indicated. (J) Sanger sequencing of the affected patients and their unaffected mother confirms the presence of a guanine to cytosine mutation leading to a charge reversing glutamic acid to glutamine substitution. This variant was identified by whole exome sequencing of the affected individuals and the unaffected mother. (K) We immunoprecipitated the FLAG tags of the wild-type and E89Q forms of CEP90 and blotted for endogenous CEP63 and FLAG.
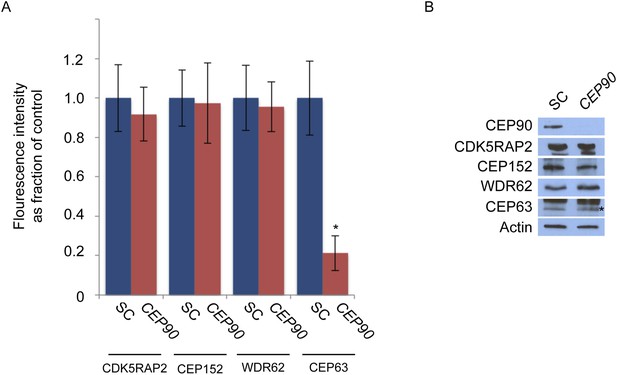
CEP90 is required to localize CEP63 to the centrosome.
(A) The centrosomal fluorescence intensity of CDK5RAP2, CEP152, WDR62 and CEP63 were quantified in SC or CEP90-depleted HeLa cells. Quantification is expressed as mean percentage ±s.d. of the fluorescence intensities of SC cells. For all quantifications 30 cells were analyzed per experiment (n = 3). p < 0.005 (paired t-test) statistically significant differences are denoted by an asterisk. (B) Total cell lysate from SC and CEP90-depleted cells were subjected to immunoblotting using antibodies to CEP90, CDK5RAP2, CEP152, WDR62, and CEP63. Actin served as a loading control.
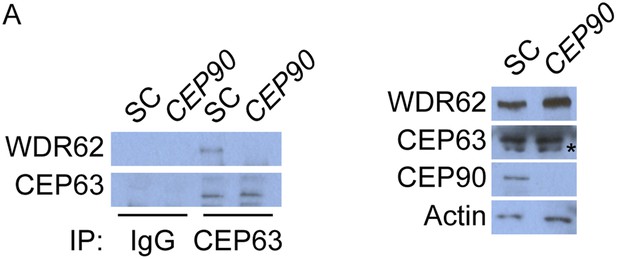
CEP63 interacts with WDR62 in a CEP90-dependent manner.
(A) HeLa total cell lysate from SC and CEP90-depleted cells were subjected to endogenous immunoprecipitation using an antibody to CEP63 and a negative control. Precipitating proteins were analyzed by immunoblotting for CEP63 and WDR62. Total cell lysates from SC and CEP90 siRNA transfected HeLa cells were analyzed by immunoblotting with antibodies to WDR62, CEP63, and CEP90. Actin served as a loading control.
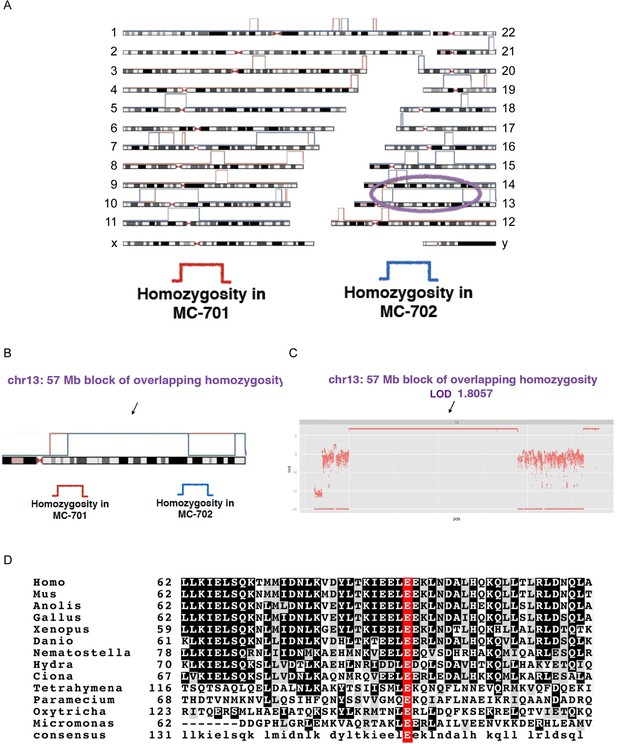
CEP90 human genetics.
(A) Schematic diagram showing linkage and homozygosity mapping in MCPH-affected individuals MC-701 and MC-702. Regions labeled with red brackets denote homozygosity in MC-701. Blue brackets denote homozygosity in MC-702. The purple oval highlights the region of shared homozygosity between the two patients. (B) 57 Mb block of overlapping homozygosity on chromosome 13, circled in (A). (C) Calculation of logarithmic odds (LOD) score over the block of overlapping homozygosity (g.chr13:30975286-88096410 in hg19 coordinates) yields a parametric, multipoint LOD score of 1.8057. (D) Phylogenetic analysis of CEP90 (using BLAST mutual best match) identified CEP90 orthologs in unicellular organisms. Analysis of the amino-terminal portion of these proteins confirms that CEP90 is ancient and that the variant affects a highly conserved residue (E89).
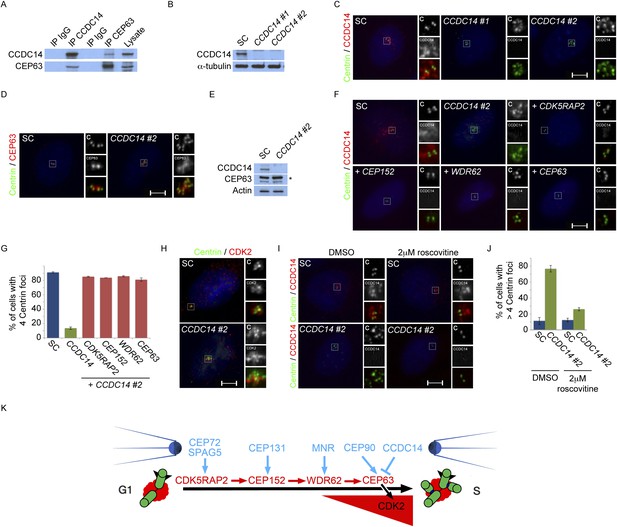
Satellite component CCDC14 suppresses the formation of supernumerary Centrin foci by limiting the centrosomal accumulation of CEP63 and activity of CDK2.
(A) We immunoprecipitated endogenous CCDC14 and CEP63 from HeLa total cell lysates. Efficient precipitation and co-precipitation were detected using antibodies specific to CCDC14 and CEP63. (B) S phase SC, CCDC14 #1, and CCDC14 #2 siRNA transfected U2OS cells were analyzed by immunofluorescence with antibodies to CCDC14 (red) and Centrin (‘c’, green). (C) Total cell lysates of SC, CCDC14 #1, and CCDC14 #2 siRNA-treated U2OS cells were analyzed by immunoblotting for CCDC14. α-tubulin served as a loading control. (D) S phase SC and CCDC14 #2 siRNA-treated U2OS cells were co-stained for Centrin (‘c’, green) and CEP63 (red). (E) Total cell lysate of SC, CCDC14 #2 siRNA transfected U2OS cells were analyzed by immunoblotting with antibodies to CCDC14 and CEP63. Actin served as a loading control. (F) U2OS cells transfected with SC siRNA, or siRNA targeting CCDC14 alone, or CCDC14 siRNA in combination with siRNAs targeting CDK5RAP2, CEP152, WDR62, or CEP63 in S phase were co-stained with Centrin (‘c’, green) and CCDC14 (red). (G) Quantification of S phase SC, CCDC14, or CCDC14 and CDK5RAP2, CEP152, WDR62, or CEP63 siRNA-treated U2OS cells with fewer than four centrioles. S phase cells were identified by Cyclin A immunostaining. For all quantifications at least 100 cells were counted per experiment (n = 3), p < 0.005 (paired t-test). (H) S phase SC and CCDC14 #2 siRNA transfected U2OS cells were co-stained with Centrin (‘c’, green) and CDK2 (red). (I) S phase SC and CCDC14 #2 siRNA transfected U2OS cells treated with DMSO or roscovitine were analyzed by immofluorescence with antibodies to CCDC14 (red) and Centrin (‘c’, green). Scale bars indicate 5 μm for all images. (J) Percentage of S phase SC, CCDC14 #2 siRNA-transfected U2OS cells treated with DMSO or roscovitine with greater than four Centrin foci. (K) CDK5RAP2 delivery to the centrosome requires the centriolar satellite proteins CEP72 and SPAG5. In a manner dependent on CDK5RAP2, CEP131 localizes CEP152 to the centrosome, MNR localizes WDR62, and CEP90 localizes CEP63. The centriolar satellite, CCDC14 binds CEP63 and removes it from the centrosome to limit centriole duplication by limiting the localization and activity of CDK2.
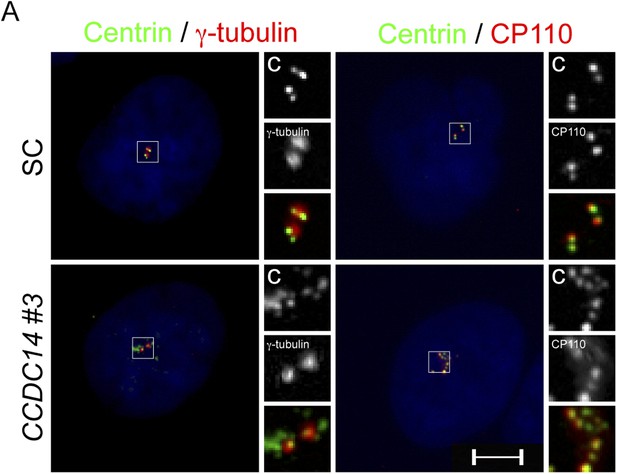
CCDC14 limits the formation of Centrin positive foci that do not recruit the PCM component γ-tubulin.
(A) Immunofluorescence images of SC and CCDC14 #2 siRNA transfected U2OS cells co-stained for Centrin (‘c’, green), γ-tubulin (red), and CP110 (red), to assess association with pericentrosomal matrix and organized centriole distal ends.
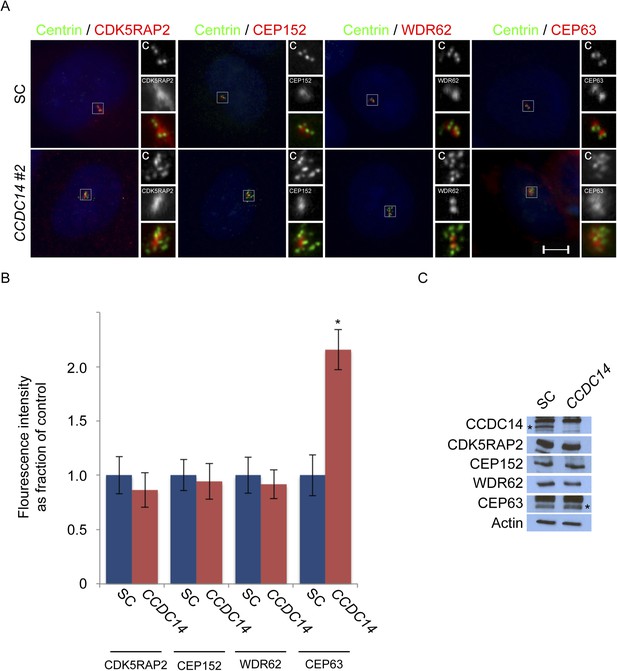
CCDC14 limits the centrosomal accumulation of CEP63 but does not alter CDK5RAP2, CEP152, or WDR62 localization.
(A) S phase SC and CCDC14 #2 siRNA transfected U2OS cells were co-stained with Centrin (‘c’, green), CDK5RAP2 (red), CEP152 (red), WDR62 (red), and CEP63 (red). (B) Quantification of the centrosomal fluorescence intensities of CDK5RAP2, CEP152, WDR62 and CEP63 in SC or CCDC14-depleted U2OS cells expressed as the mean percentage ±s.d. of the fluorescence intensities of SC cells. For all quantifications 30 cells were analyzed per experiment (n = 3). p < 0.005 (paired t-test) statistically significant differences are denoted by an asterisk. (C) Total cell lysate from SC and CCDC14-depleted HeLa cells was subjected to immunoblotting using antibodies to CCDC14, CDK5RAP2, CEP152, WDR62 and CEP63. Actin served as a loading control.
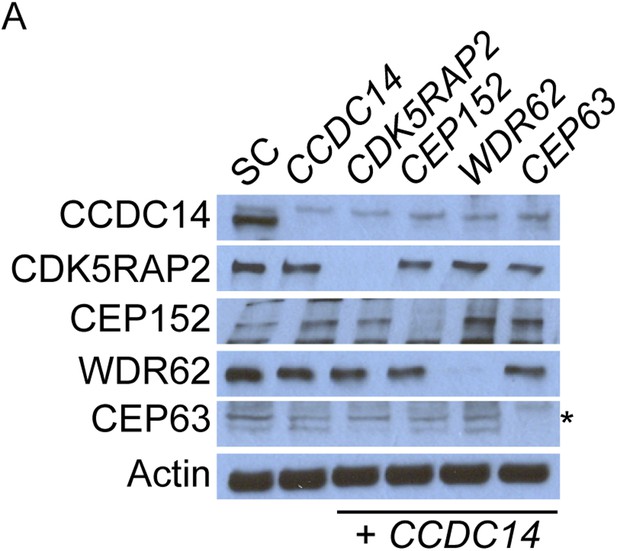
Co-depletion of CCDC14 and MCPH-associated proteins.
(A) Total cell lysate of SC, CCDC14, or CCDC14 plus CDK5RAP2, CEP152, WDR62, or CEP63 siRNA transfected U2OS cells were analyzed by immunoblotting with antibodies to CCDC14, CDK5RAP2, CEP152, WDR62, and CEP63. Actin served as a loading control. Asterisk indicates the specific band.
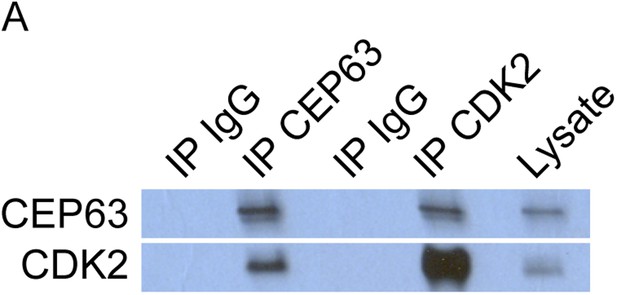
CEP63 and CDK2 interact.
(A) HeLa total cell lysate was subjected to immunoprecipitation of using antibodies to endogenous CDK2, CEP63 and a negative control, FLAG. Precipitating proteins were analyzed by immunoblotting for CEP63 and CDK2.
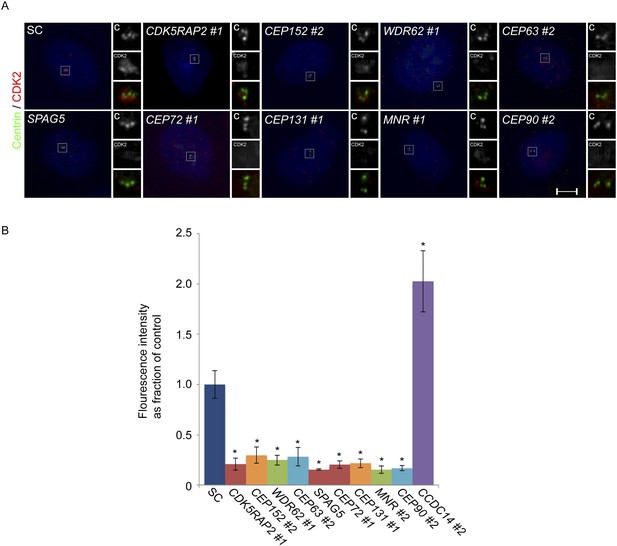
CDK2 localizes to the centrosome in a CDK5RAP2, CEP152, WDR62, CEP63, SPAG5, CEP72, CEP131, MNR, and CEP90-dependent manner.
(A) S phase SC, CDK5RAP2, CEP152, WDR62, CEP63, SPAG5, CEP72, CEP131, MNR, and CEP90 siRNA transfected U2OS cells were analyzed by immunofluorescence with antibodies to Centrin (‘c’, green) and CDK2 (red). (B) Quantification of the mean fluorescence intensities ±s.d. of CDK2 in SC, CDK5RAP2, CEP152, WDR62, CEP63, SPAG5, CEP72, CEP131, MNR, CEP90 and CCDC14 siRNA treated cells expressed as the mean percentage ±s.d. of the fluorescence intensities of SC cells. For all quantifications 30 cells were analyzed per experiment (n = 3). p < 0.005 (paired t-test) statistically significant differences from SC controls are denoted by an asterisk.
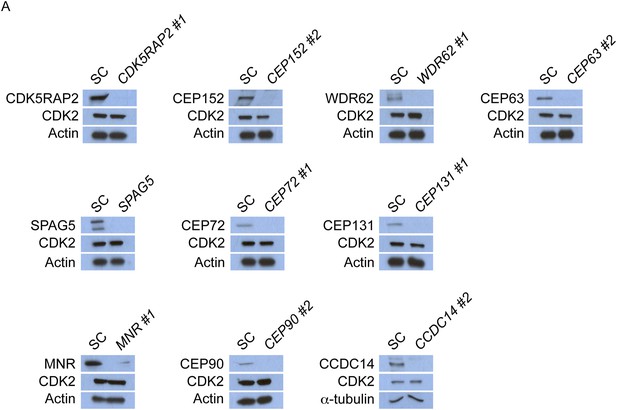
CDK5RAP2, CEP152, WDR62, CEP63, SPAG5, CEP72, CEP131, MNR, CEP90 and CCDC14 do not control the stability of CDK2.
(A) Total cell lysate of SC, CDK5RAP2, CEP152, WDR62, CEP63, SPAG5, CEP72, CEP131, MNR, CEP90 and CCDC14 siRNA transfected HeLa cells were analyzed by immunoblotting with antibodies to CDK5RAP2, CEP152, WDR62, CEP63, SPAG5, CEP72, CEP131, MNR, CEP90, CCDC14 and CDK2. Actin or α-tubulin served as a loading control.
Additional files
-
Supplementary file 1
CDK5RAP2, CEP152, WDR62 and CEP63 mass spectrometry analysis. (A–D) Selected list of CDK5RAP2, CEP152, WDR62, and CEP63 interacting centrosomal proteins, including peptide counts and percent coverage. Coprecipitating proteins were counterscreened against c-Myc and FLAG interacting proteins, which served as negative controls. We used the proteomic analysis by Andersen et al. (2003) and Jakobsen et al. (2011) to suggest the identity of centrosomal proteins for additional analysis. Precipitated proteins are highlighted in blue, MCPH-associated proteins are in green, and centriolar satellite proteins are in orange.
- https://doi.org/10.7554/eLife.07519.035
-
Supplementary file 2
siRNA sequences. Human Stealth siRNAs to CDK5RAP2, WDR62, CEP63, SPAG5, CEP72, CEP131, MNR, CEP90 and CCDC14 were obtained from Life Technologies. SC, CEP152, CEP63 NB and CEP215 siRNAs were synthesized by Life Technologies.
- https://doi.org/10.7554/eLife.07519.036





