The Arabidopsis active demethylase ROS1 cis-regulates defence genes by erasing DNA methylation at promoter-regulatory regions
Figures
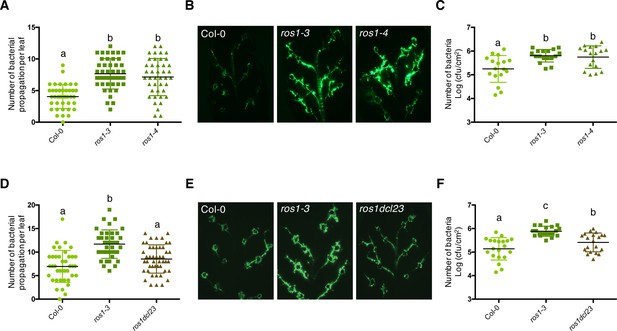
The enhanced Pto DC3000 disease susceptibility observed in ros1-infected mutants is mainly dependent on DCL2 and/or DCL3 functions.
(A) Increased Pto DC3000 vascular propagation in the two independent ros1 mutant alleles, ros1-3 and ros1-4. Secondary veins of 5-week-old Col-0, ros1-3 and ros1-4 mutants were inoculated with a virulent GFP-tagged Pto DC3000 strain (Pto DC3000-GFP) at 5 × 106 cfu ml−1 using the toothpick inoculation method. Inoculation was done on six secondary veins per leaf and two sites of inoculation per vein. At least 10 leaves per experiment were quantified. The number of Pto DC3000-GFP spreading events from the wound inoculation sites was quantified after 3 days under UV light using a macrozoom. When the bacteria propagated away from any of the 12 inoculation sites, it was indexed as propagation with a possibility of maximum 18 propagations per leaf. The values from three independent experiments were considered for the comparative analysis. Statistical significance was assessed using a one-way ANOVA test and Tukey’s multiple comparisons test. (B) Representative pictures of the GFP fluorescence observed at the whole leaf level on the plants depicted in A. (C) Enhanced Pto DC3000 apoplastic growth in the two independent ros1 mutant alleles, ros1-3 and ros1-4. Five-week-old plants of Col-0, ros1-3, and ros1-4 mutants were dip-inoculated with Pto DC3000-GFP at 5 × 107 cfu ml−1. Bacterial titres were monitored at 3 days post-inoculation (dpi). Each data point represents bacterial titre at four leaf discs extracted from a single leaf. Two leaves out of three plants per line per experiment, and from three independent experiments were considered for the comparative analysis. Statistical significance was assessed using a one-way ANOVA test and Tukey’s multiple comparisons test. (D) Increased bacterial propagation in the vein observed in ros1-3 is rescued in the ros1dcl23 triple mutant. Secondary veins of 5-week-old Col-0, ros1-3 and the triple ros1dcl23 mutants were inoculated as in A and the results analysed as in A. (E) Representative pictures of the GFP fluorescence observed at the whole leaf level on the plants presented in D. (F) Enhanced Pto DC3000 apoplastic growth in ros1 is partially rescued in the ros1dcl23 background. Five-week-old plants of Col-0, ros1-3, and ros1dcl23 were inoculated as in C and the results were analysed as in C.
-
Figure 1—source data 1
Original data of bacterial propagation assays for Figure 1A,C,D, and F.
- https://cdn.elifesciences.org/articles/62994/elife-62994-fig1-data1-v2.xlsx
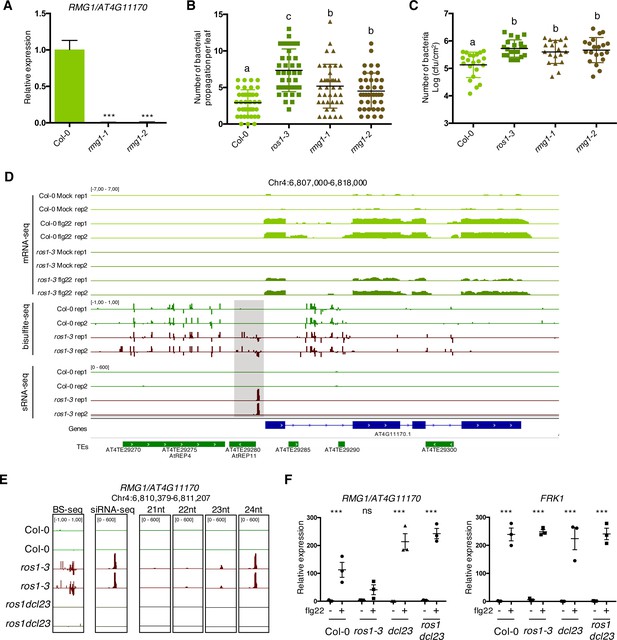
The TNL gene RMG1 contributes to apoplastic and vascular resistance against Pto DC3000 and its flg22-triggered induction is negatively regulated by RNA-directed DNA methylation (RdDM) in the absence of ROS1.
(A) RMG1 mRNA levels in Col-0 and the two rmg1 mutant alleles, namely rmg1-1 and rmg1-2, were monitored by RT-qPCR at 6 hr after syringe-infiltration of mock (water) or 1 μM of flg22 peptide. (B) RMG1 positively regulates vascular resistance towards Pto DC3000. Secondary veins of 5-week-old Col-0, ros1-3, rmg1-1, and rmg1-2 plants were inoculated with Pto DC3000-GFP at 5 × 106 cfu ml−1 using the toothpick inoculation method. Inoculation was done on six secondary veins per leaf and two sites of inoculation per vein. At least 10 leaves per condition were quantified. The number of Pto DC3000-GFP spreading events from the wound inoculation sites was quantified after 3 days under UV light using a macrozoom. When the Pto DC3000-GFP propagated away from any of the 12 inoculation sites, it was indexed as propagation with a possibility of maximum 18 propagations per leaf. The values from three independent experiments were considered for the comparative analysis. Statistical significance was assessed using a one-way ANOVA test. (C) RMG1 positively regulates apoplastic resistance towards Pto DC3000. Five-week-old Col-0, ros1-3, rmg1-1, and rmg1-2 plants were dip-inoculated with Pto DC3000-GFP at 5 × 107 cfu ml−1. Bacterial titres were monitored at 3 days post-infection (dpi). Each data point represents bacterial titre at four leaf discs extracted from two different leaves. At least three leaves out of four plants per line per experiment and from three independent experiments were considered for the comparative analysis. Statistical significance was assessed using a one-way ANOVA test. (D) Flg22-triggered induction of RMG1 is compromised in ros1-3-elicited mutant and correlates with an increased DNA methylation and siRNA levels at the remnant RC/Helitron TE AtREP11, and particularly at its 3’ boundary. IGV snapshots showing mRNA levels (mRNA-seq) after syringe-infiltration of mock (water) or 1 μM of flg22 peptide for Col-0 and ros1-3, and cytosine DNA methylation levels (Bs-Seq) and siRNA levels (sRNA-seq) in 5-week-old untreated rosette leaves of Col-0 and ros1-3, at the RMG1 locus. The hyperDMR is highlighted by the dotted box. (E) Levels of different siRNA species at the 3’ boundary of AtREP11. IGV snapshots representing the levels of methylation (BS-seq), total siRNA species (siRNA-seq), and different size of siRNA species (21nt, 22nt, 23nt, and 24nt siRNAs) in 5-week-old rosette leaves of Col-0, ros1-3, and ros1dcl23. (F) The flg22-triggered induction of RMG1 is fully restored in ros1dcl23-elicited triple mutants. RT-qPCR analysis depicting RMG1 and FRK1 mRNA levels in Col-0, ros1-3, dcl23, and ros1dcl23 5-week-old rosette leaves treated with either mock (water) or 1 μM of flg22 for 6 hr. The mRNA levels are relative to the level of UBQ transcripts. Statistical significance of flg22 treatment on expression was assessed using a two-way ANOVA test and a Sidak’s multiple comparisons test. Asterisks indicate statistical significance (*: p<0.05, **: p<0.01, ***: p<0.001, ns: not significant).
-
Figure 2—source data 1
Original qRT-PCR data for Figure 2A and F, and bacterial propagation data for Figure 2B and C.
- https://cdn.elifesciences.org/articles/62994/elife-62994-fig2-data1-v2.xlsx
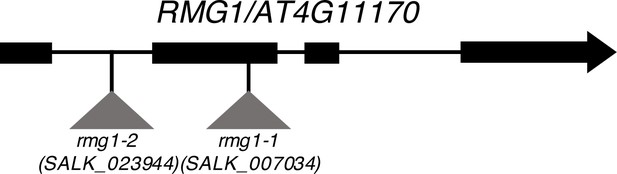
Scheme representing T-DNA insertion sites at the RMG1 locus.
The T-DNA insertions SALK_007034 (rmg1-1 allele) and SALK_023944 (rmg1-2 allele) are located in the second exon and in the first intron of RMG1 (AT4G11170), respectively.
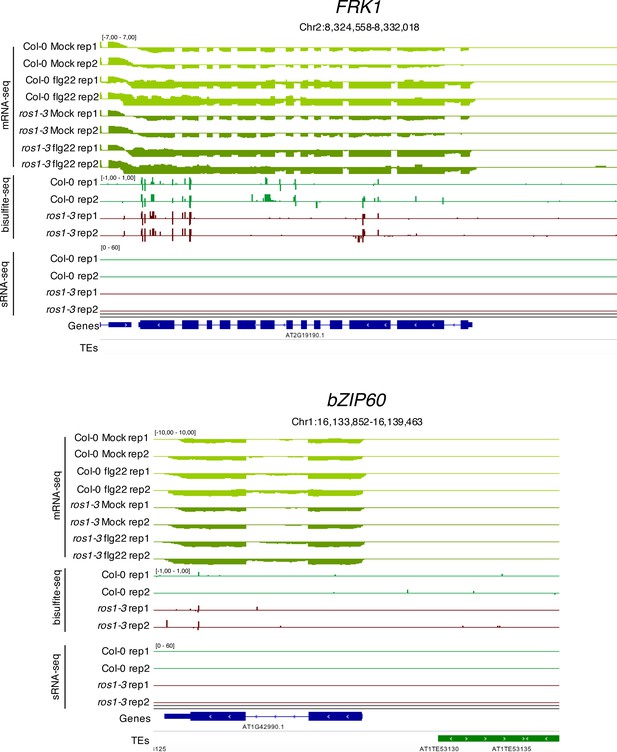
FRK1 and bZIP60 promoters are not hypermethylated in ros1-3 mutants.
IGV snapshots showing mRNA levels (mRNA-seq) after syringe-infiltration of mock (water) or 1 μM of flg22 peptide in 5-week-old rosette leaves of Col-0 and ros1-3, and cytosine methylation levels (Bs-Seq) and siRNA levels (sRNA-seq) in 5-week-old untreated rosette leaves of Col-0 and ros1-3, at the Flg22-induced Receptor-like Kinase 1 (FRK1) (upper panel) and bZIP60 (lower panel) loci.
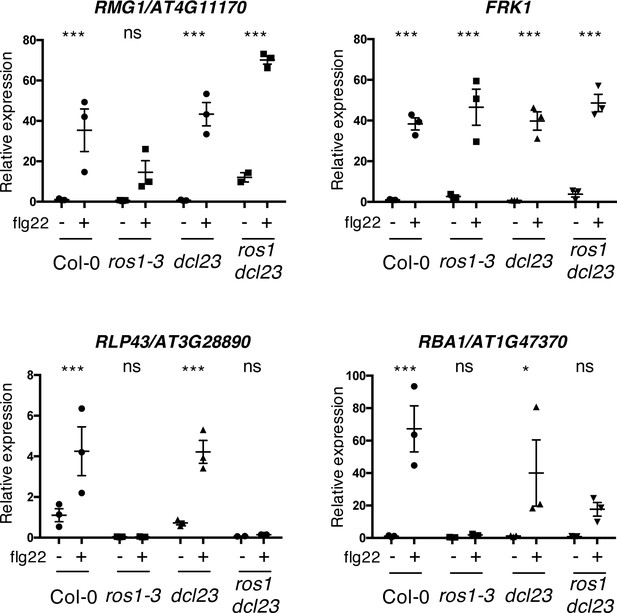
The flg22-triggered induction of RMG1 and RBA1 is restored, while RLP43 remains in a repressed state, in the ros1dcl23-elicited mutant.
Second biological replicate of RT-qPCR analyses depicting RMG1, FRK1, RLP43, and RBA1 mRNA levels in 5-week-old rosette leaves of Col-0, ros1-3, dcl23, and ros1dcl23 treated with either mock (water) or 1 μM of flg22 for 6 hr. The mRNA levels are relative to the level of UBQ transcripts. Statistical significance of flg22 treatment on expression was assessed using a two-way ANOVA test and a Sidak’s multiple comparisons test. Asterisks indicate statistical significance (*: p<0.05, **: p<0.01, ***: p<0.001, ns: not significant).
-
Figure 2—figure supplement 3—source data 1
Original qRT-PCR data for Figure 2—figure supplement 3.
- https://cdn.elifesciences.org/articles/62994/elife-62994-fig2-figsupp3-data1-v2.xlsx
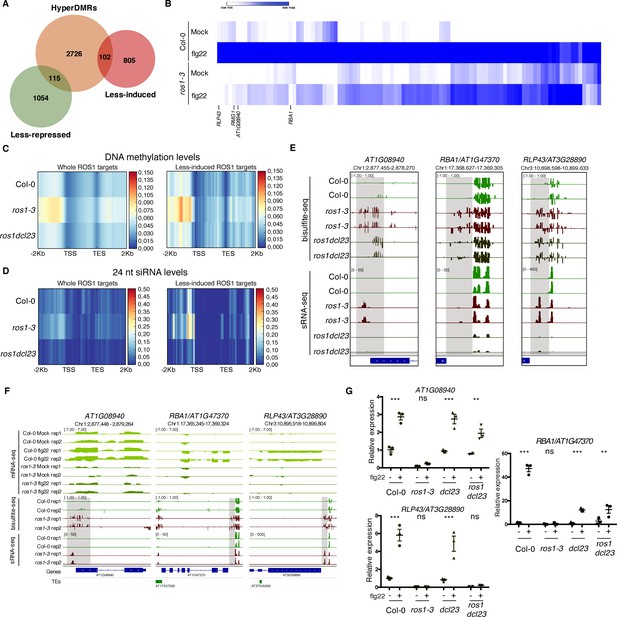
Genome-wide identification of flg22-responsive ROS1 targets and characterisation of the role of RNA-directed DNA methylation (RdDM) in the methylation status of these genes in the absence of ROS1.
(A) Proportion of flg22-responsive genes that are regulated by ROS1. One hundred and two flg22-responsive genes that are ‘less-induced’ and 115 flg22-responsive genes that are ‘less-repressed’ in ros1-3-elicited mutant exhibit hypermethylated DMRs (hyperDMRs). Venn diagram representing the overlap of genes presenting hyperDMRs in the ros1 mutant (in orange) with genes presenting a compromised induction (in red) or repression (in green) in ros1-3 compared to Col-0 treated with mock (water) or 1 μM of flg22 for 6 hr. (B) Heat map representing the relative expression of the 102 less-induced genes in Col-0 and ros1-3 treated with mock (water) or 1 μM of flg22 for 6 hr. Merged data from two independent biological replicates are presented. (C) Increased global DNA methylation levels observed in untreated ros1-3 compared to Col-0 at the whole set of genes exhibiting hyperDMRs (left panel) and at the 102 less-induced genes (right panel) are restored in the ros1dcl23 triple mutant. Heatmap representing global DNA methylation levels within regions comprising 2 Kb upstream of the transcription start site (TSS), the gene body, and 2 Kb downstream of the transcription end site (TES) of the whole set of genes exhibiting hyperDMRs in ros1-3 versus Col-0 (left panel) and of the 102 less-induced genes (right panel). These heatmaps were generated from BS-seq data sets obtained from 5-week-old rosette leaves of Col-0, ros1-3, and ros1dcl23 mutants. (D) Increased 24 nt siRNA levels in ros1-3 at the whole set of genes exhibiting hyperDMRs in ros1-3 versus Col-0 (left panel) and at the 102 less-induced genes (right panel) are restored in the ros1dcl23 triple mutant. Heatmap representing 24-nt siRNA levels within regions comprising 2 Kb upstream of the TSS, the gene body, and 2 Kb downstream of the TES of the whole set of genes exhibiting hyperDMRs in ros1-3 versus Col-0 (left panel) and of the 102 less-induced genes (right panel). These heatmaps were generated from sRNA-seq datasets obtained from 5-week-old rosette leaves of untreated Col-0, ros1-3 and ros1dcl23 mutants. Average of the two replicates is represented. (E) Methylation levels are restored at AT1G08940 and partially restored at RBA1 in ros1dcl23 whereas RLP43 retains methylation levels similar to methylation levels observed in the single ros1-3 mutant. IGV snapshots showing siRNA levels and methylation levels at the DMRs of AT1G08940, RBA1, and RLP43 in Col-0, ros1-3, and ros1dcl23. (F) IGV snapshots depicting mRNA-seq data in Col-0 and ros1-3 mutant in mock- and flg22-treated conditions as well as BS-seq and sRNA-seq of untreated Col-0 and ros1-3 plants. (G) AT1G08940 gene induction is restored and RBA1 gene induction is partially restored whereas RLP43 remains in a repressed state in ros1dcl23. RT-qPCR analyses from 5-week-old rosette leaves of Col-0, ros1-3, dcl23, and ros1dcl23 treated with either mock (water) or 1 μM of flg22 for 6 hr. The mRNA levels are relative to the level of UBQ transcripts. Statistical significance of flg22 treatment on expression was assessed using a two-way ANOVA test and a Sidak’s multiple comparisons test. Asterisks indicate statistical significance (*: p<0.05, **: p<0.01, ***: p<0.001, ns: not significant).
-
Figure 3—source data 1
Original qRT-PCR data for Figure 3G.
- https://cdn.elifesciences.org/articles/62994/elife-62994-fig3-data1-v2.xlsx
-
Figure 3—source data 2
Original transcriptomic data used for the heatmap presented in Figure 3B.
- https://cdn.elifesciences.org/articles/62994/elife-62994-fig3-data2-v2.xlsx
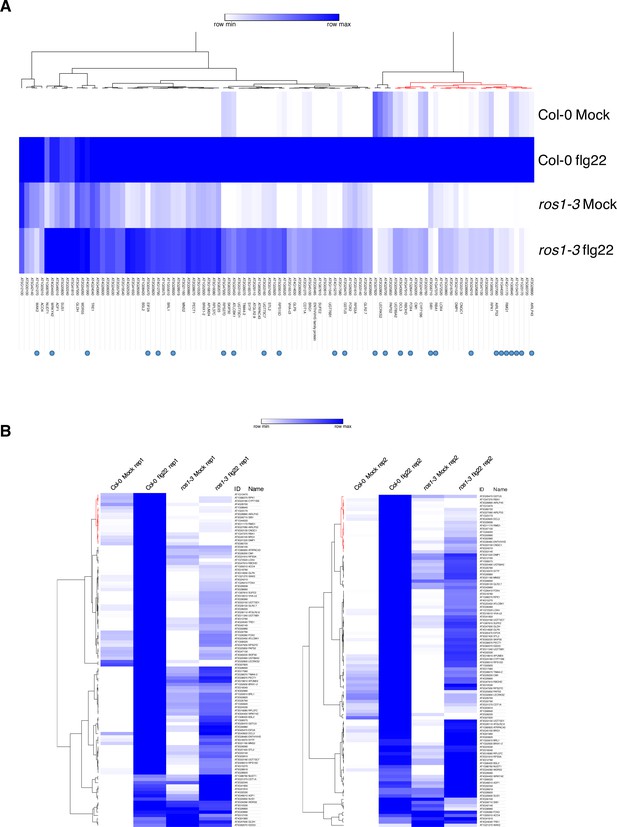
RNA sequencing experiment in 5-week-old rosette leaves of Col-0 and ros1-3 syringe-infiltrated with either mock or flg22 for 6 hr.
(A) Heatmap depicting the relative expression of the 102 less-induced genes in Col-0 and ros1-3 mutants upon 6 hr of mock or flg22 treatments. AGI numbers and gene names are shown for all individual gene. Merged data from two independent biological replicates are presented. (B) Heatmaps showing the relative expression of the 102 less-induced genes for each independent biological replicate.
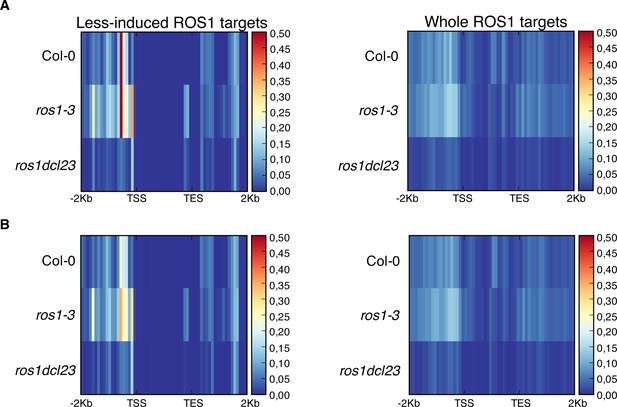
Analysis of 23–24 nt siRNA levels for the groups of 102 less-induced genes and 2943 genes carrying hyperDMRs in the ros1-3 mutant background.
(A) Heatmaps depicting 23nt- and 24nt-long siRNA levels at the 102 less-induced genes (left panel) and at the 2943 genes presenting hyperDMR in ros1-3 (right panel) on replicate 1. Global siRNA levels were quantified from 2 Kb upstream sequence from the TSS, the gene body, and 2 Kb downstream sequence of the TES of genes in 5-week-old rosette leaves in non-treated Col-0, ros1-3, and ros1dcl23. (B) Heatmaps depicting 23nt- and 24nt-long siRNA levels at the 102 less-induced genes (left panel) and at the 2943 genes presenting hyperDMR in ros1-3 (right panel) on replicate 2. Global siRNA levels were quantified from 2 Kb upstream sequence from the transcription start site (TSS), the gene body, and 2 Kb downstream sequence of the TES of genes in 5-week-old rosette leaves in non-treated Col-0, ros1-3, and ros1dcl23.
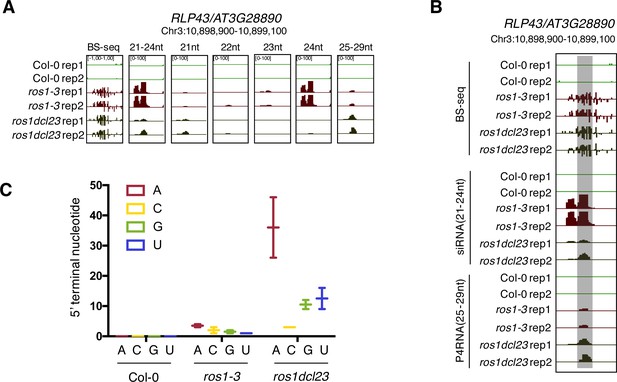
Gain of 21-nt siRNAs and P4RNAs at the RLP43 promoter in the ros1dcl23 background might contribute to the maintenance of DNA methylation levels in this triple mutant.
(A) Snapshot representing levels of DNA methylation and siRNA species classified per size at the RLP43 promoter, in 5-week-old rosette leaves of untreated Col-0, ros1-3, and ros1dcl23. (B) Snapshot representing levels of DNA methylation, 21–24 nt siRNAs and 25–29 nt P4RNAs at the RLP43 promoter, in 5-week-old rosette leaves of untreated Col-0, ros1-3, and ros1dcl23. The region presenting increased P4RNAs is highlighted in the grey box. (C) Number of putative P4RNAs classified depending on the nucleotide present at the 5’ terminal in Col-0, ros1-3, and ros1dcl23. Data from the two biological replicates are presented.
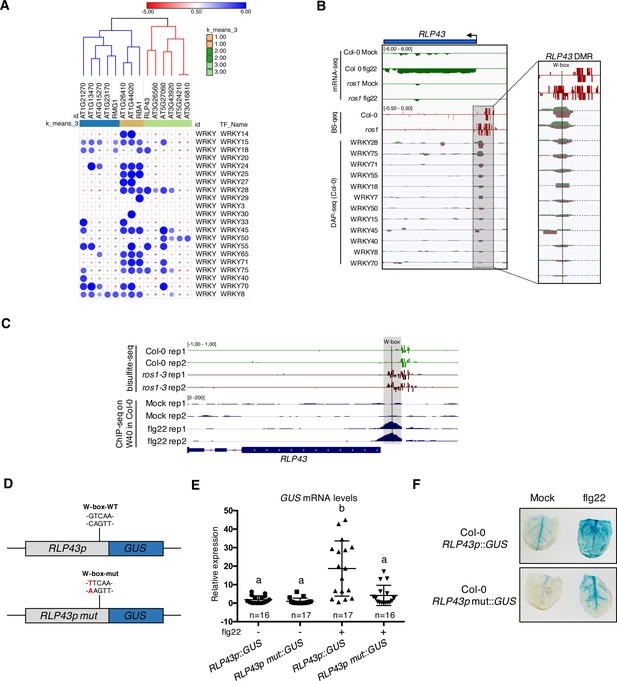
Several WRKY transcription factors bind to the demethylated region of the RLP43 promoter, which contains a functional and flg22-responsive W-box cis-element.
(A) A subgroup of the ROS1 targets exhibits an over-representation of WRKY DNA binding at the promoter regions corresponding to the hyperDMRs that were retrieved in ros1-3. GAT analysis performed on publicly available DNA affinity purification sequencing (DAP-seq) data (O'Malley et al., 2016) identified WRKY TFs with significant enrichment in the regions corresponding to the hyperDMRs observed in ros1-3 for the 14 stringent ROS1 primary targets. (B) Several WRKY TFs specifically bind to the region that is demethylated by ROS1 in the RLP43 promoter. Snapshots representing, from top to bottom, mRNA-seq (Rep1), BS-seq (Rep1), and DAP-seq data (O'Malley et al., 2016) at the RLP43 locus. To better appreciate the overlap between the promoter region of RLP43 subjected to ROS1-directed demethylation and the region where WRKY TFs bind to DNA, a zoom in is depicted at the level of the hyperDMR (box on the right panel) and the position of the W-box is highlighted by the vertical dashed line. (C) A hemagglutinin (HA) epitope-tagged version of WRKY40 binds in vivo to the ROS1-targeted region of the RLP43 promoter in a flg22-dependent manner. Snapshots depicting bisulfite sequencing data from 5-week-old rosette leaves of Col-0 and ros1-3 mutant (Rep1) and ChIP-seq data performed on seedlings of wrky40 mutants (SLAT collection of dSpm insertion line; Shen et al., 2007) complemented with WRKY40-HA treated with either mock (medium without flg22) or flg22 (medium supplemented with flg22) for 2 hr, at RLP43 (Birkenbihl et al., 2017). (D) Scheme representing the RLP43 transcriptional GUS fusion constructs containing the WT W-box sequence (RLP43p::GUS) and the mutated W-box sequence (RLP43p mut::GUS). (E) The W-box cis-element at the RLP43 promoter is functional and responsive to flg22. RT-qPCR analyses were performed to monitor the GUS mRNA levels in primary T1 transformants expressing either the RLP43p::GUS or RLP43p mut::GUS transgenes. For each individual, two leaves were syringe-infiltrated with mock (water) and two other leaves were treated the same way with flg22 at a concentration of 1 μM for 6 hr. The GUS mRNA levels are relative to the level of UBQ transcripts. Statistical significance was assessed using a one-way ANOVA test and a Tukey’s multiple comparisons test. (F) The flg22-induced GUS activity is impaired in RLP43p mut::GUS-elicited plants. GUS-staining of 5-week-old rosette leaves of RLP43p::GUS or RLP43pmut::GUS primary transformants that were syringe-infiltrated with either mock or 1 μM of flg22 for 24 hr. Representative pictures are shown.
-
Figure 4—source data 1
Original qRT-PCR data of GUS transcript quantification for Figure 4E.
- https://cdn.elifesciences.org/articles/62994/elife-62994-fig4-data1-v2.xlsx

GAT analysis of all the transcription factor classes binding to the 26 stringent less-induced ROS1 target promoters.
(A) GAT analysis performed on publicly available DNA affinity purification sequencing (DAP-seq) data (O'Malley et al., 2016) for WRKY TFs at the 26 stringent ROS1 primary targets. This analysis was conducted on 26 stringent less-induced ROS1 targets, from which promoter hyperDMR sequences exhibiting dense methylated levels in ros1-3 mutants were manually selected (these genes are represented by blue dots in Figure 4—figure supplement 4). (B) GAT analysis performed on publicly available DAP-seq data (O'Malley et al., 2016) identified TFs with significant enrichment at the promoter demethylated regions from the 26 stringent less-induced ROS1 targets.
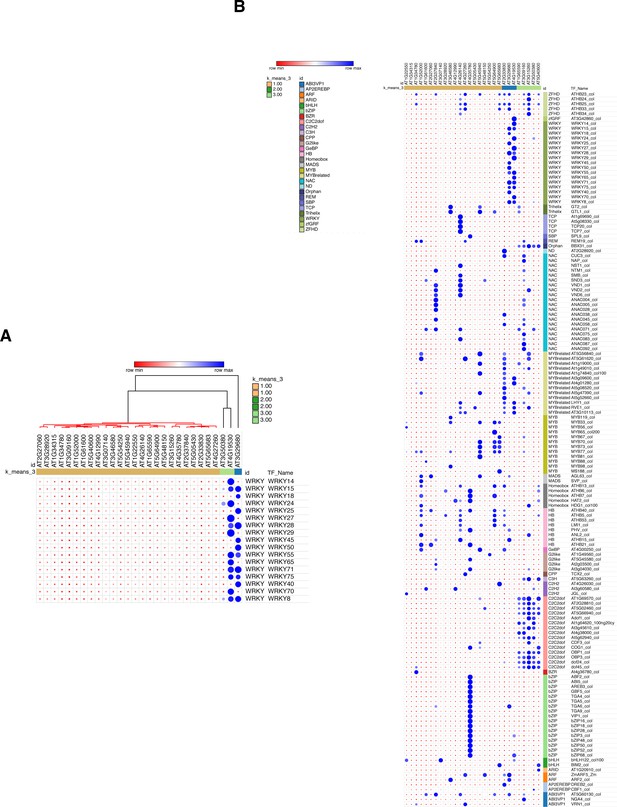
GAT analysis of all the transcription factor classes binding to the 28 stringent less-repressed ROS1 target promoters.
(A) GAT analysis performed on publicly available DNA affinity purification sequencing (DAP-seq) data (O'Malley et al., 2016) for WRKY TFs at the 28 stringent less-repressed ROS1 targets, from which promoter hyperDMR sequences exhibiting dense methylated levels in ros1-3 mutants were manually selected. (B) GAT analysis performed on publicly available DAP-seq data (O'Malley et al., 2016) identified TFs with significant enrichment at the promoter demethylated regions from the 28 stringent less-repressed ROS1 targets.
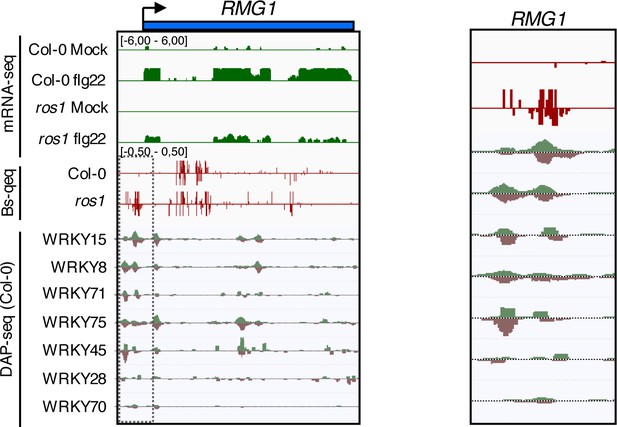
WRKY DNA binding peaks are present at the RMG1 promoter region that is subjected to ROS1-directed demethylation.
Snapshots representing, from top to bottom, RNA-seq, BS-seq, and DNA affinity purification sequencing (DAP-seq) data sets at the RMG1 locus (left panel) with an enlargement surrounding the hyperDMR and the WRKY-binding peaks (right panel).
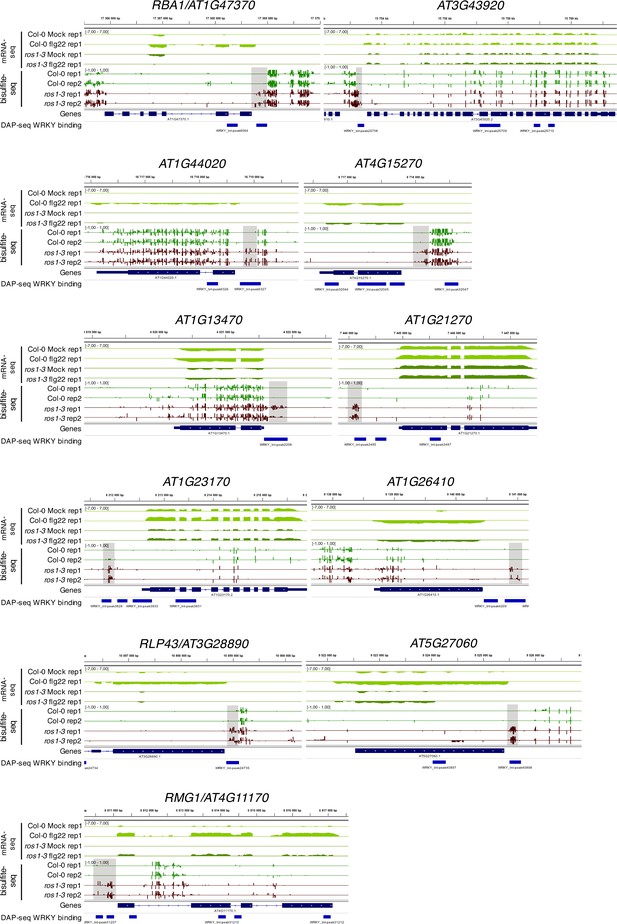
Position of WRKY DNA binding peaks at the 11 flg22-induced ROS1 targets.
Snapshots depicting RNA-seq (rep1), BS-seq (rep1 and rep2), and the position of WRKY-binding sites observed in the DNA affinity purification sequencing (DAP-seq) data (O'Malley et al., 2016) in Col-0 and ros1-3 untreated rosette leaves.
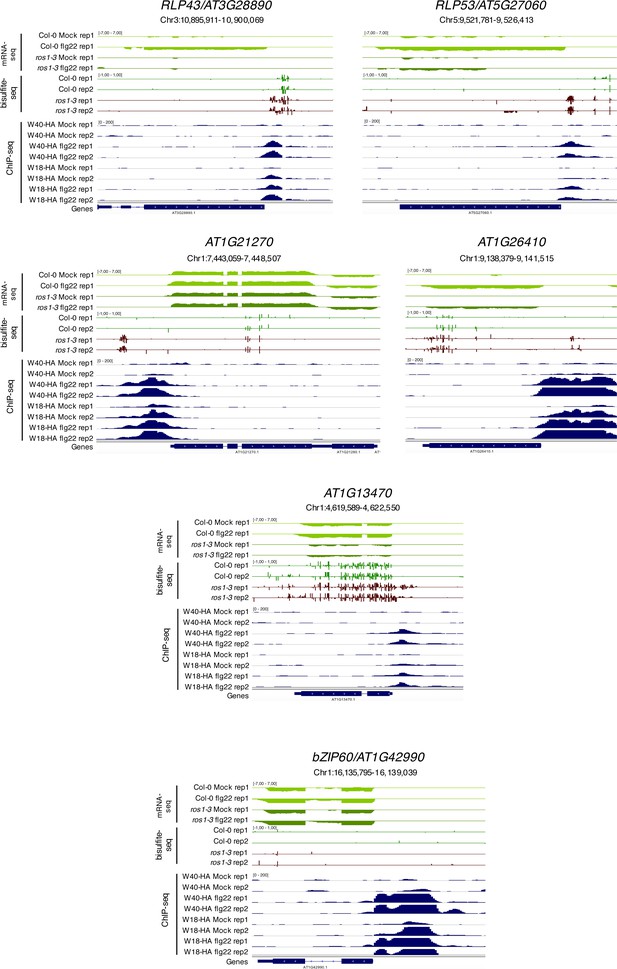
Flg22-triggered chromatin association of WRKY18-HA and WRKY40-HA at the promoters of five less-induced ROS1 targets and at the promoter of the positive control bZIP60.
Snapshots representing mRNA-seq and BS-seq of Col-0 and ros1 untreated rosette leaves and ChIP-seq data for wrky18 WRKY18-HA and wrky40 WRKY40-HA seedlings subjected to either mock (medium without flg22) or flg22 (medium supplemented with flg22) for 2 hr, at RLP43, RLP53/AT5G27060, AT1G21270, AT1G26410, AT1G13470, and bZIP60 (Birkenbihl et al., 2017).
ROS1-directed demethylation is crucial for the DNA binding of WRKY18 and WRKY40 at the RLP43 promoter.
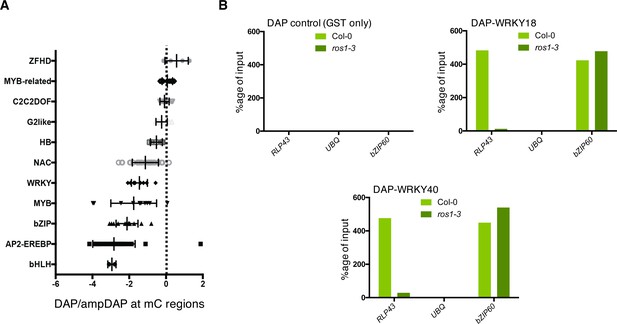
(A) DAP-seq data from O'Malley et al., 2016 showing that WRKY family members are generally sensitive to DNA methylation. Graph representing ratio of binding capacity in DAP versus ampDAP data at regions methylated in Col-0 and for the different transcription factor family members used in this study. (B) The ability of WRKY18 and WRKY40 to bind DNA corresponding to the demethylated region of the RLP43 promoter is abolished in the ros1-3 mutant background. DAP-qPCR analysis at the RLP43 promoter upon pull-down of Col-0 or ros1-3 genomic DNA by GST (negative control), WRKY18-GST or WRKY40-GST. UBQ and bZIP60 served as negative and positive controls, respectively.
-
Figure 5—source data 1
Original DAP-qPCR data for Figure 5B.
- https://cdn.elifesciences.org/articles/62994/elife-62994-fig5-data1-v2.xlsx
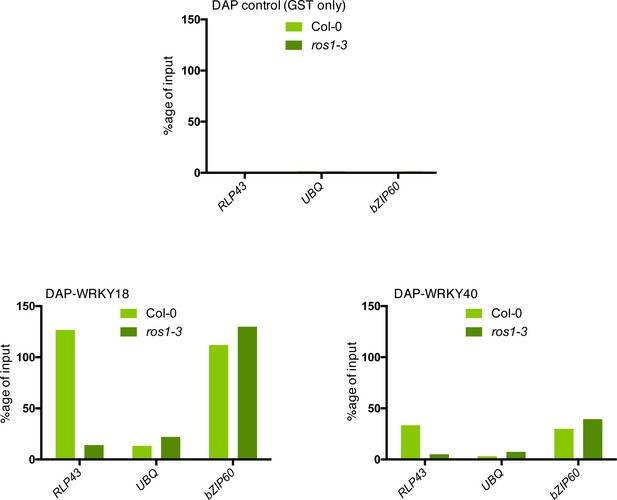
ROS1-directed demethylation is crucial for the binding of WRKY18 and WRKY40 at the RLP43 promoter.
The ability of WRKY18 and WRKY40 to bind DNA corresponding to the demethylated region of the RLP43 promoter is abolished in the ros1-3 mutant background. Second biological replicate of the DAP-qPCR analysis at the RLP43 promoter upon pull-down of Col-0 or ros1-3 genomic DNA by GST (negative control) (upper panel), WRKY18-GST (left panel), or WRKY40-GST (right panel). UBQ and bZIP60 served as negative and positive controls, respectively.
-
Figure 5—figure supplement 1—source data 1
Original DAP-qPCR data for Figure 5—figure supplement 1.
- https://cdn.elifesciences.org/articles/62994/elife-62994-fig5-figsupp1-data1-v2.xlsx
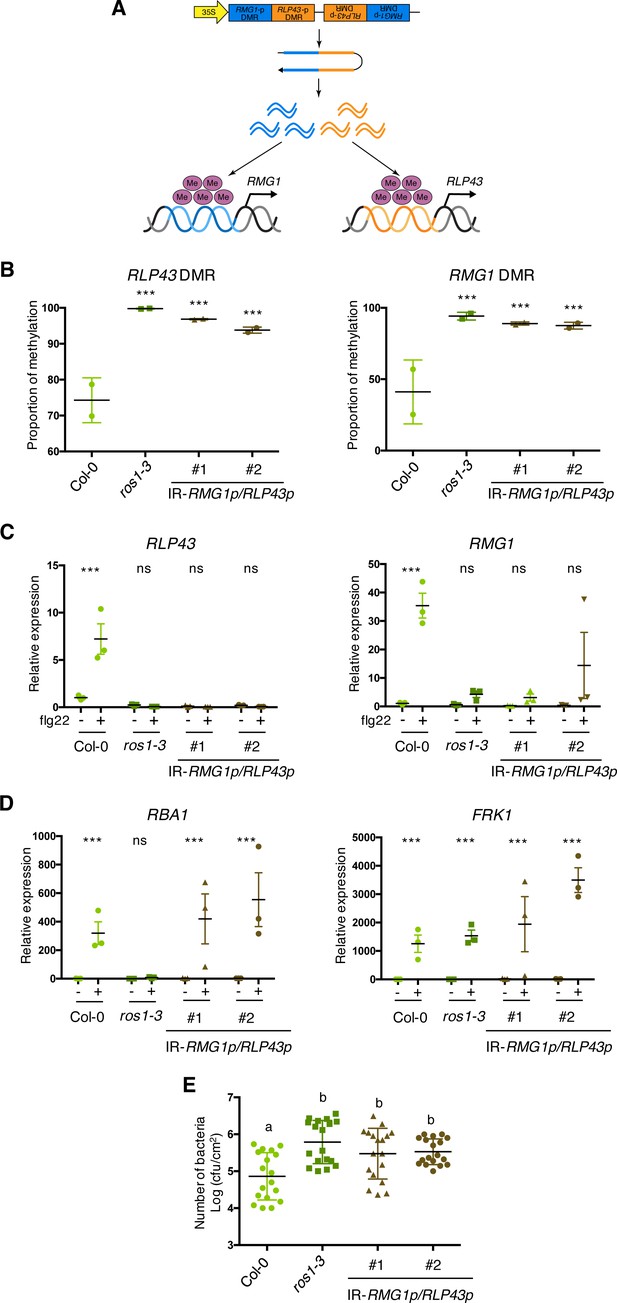
The artificial siRNA-directed targeting of remethylation at the RMG1 and RLP43 promoters impairs the flg22-triggered inducibility of these genes and enhances susceptibility towards Pto DC3000.
(A) Scheme depicting the chimeric inverted repeat (IR) construct designed to simultaneously direct DNA remethylation at the RMG1 and RLP43 promoter regions. The IR-RMG1p/RLP43p contains the sequences corresponding exactly to the promoter sequence regions of RMG1 (blue) and RLP43 (orange) that are subjected to ROS1-directed demethylation in Col-0 and hypermethylated in ros1 mutants. This inverted repeat transgene is driven by the constitutive 35S promoter, hence hypothesized to constitutively produce two populations of siRNA species designed to force remethylation of the RMG1 and RLP43 promoter regions that are normally demethylated by ROS1 in Col-0. (B) The RMG1 and RLP43 promoters exhibit hypermethylation in IR-RMG1p/RLP43p lines such as in ros1-3 mutants. Genomic DNAs from Col-0, ros1-3, and two independent IR-RMG1p/RLP43p lines (two biological replicates per line) were digested using McrBC and further analysed by qPCR. Ratio between digested DNA and undigested DNA was quantified to assess the proportion of methylation. (C) The flg22-triggered induction of RMG1 and RLP43 is impaired in the two independent IR-RMG1p/RLP43p lines. Five-week old rosette leaves of Col-0, ros1-3, and two independent IR-RMG1p/RLP43p lines were syringe-infiltrated with either mock (water) or 1 μM of flg22 for 6 hr, and the mRNA levels of RMG1 and RLP43 were monitored by RT-qPCR analyses. The mRNA levels are relative to the level of UBQ transcripts. Statistical significance of flg22 treatment on expression was assessed using a two-way ANOVA test and a Sidak’s multiple comparisons test. Asterisks indicate statistical significance (*: p<0.05, **: p<0.01, ***: p<0.001, ns: not significant). (D) The flg22-triggered induction of RBA1 and FRK1 is not affected in the two independent IR-RMG1p/RLP43p lines. Five-week old rosette leaves of Col-0, ros1-3, and two independent IR-RMG1p/RLP43p lines were syringe-infiltrated with either mock (water) or 1 μM of flg22 for 6 hr, and the mRNA levels of RBA1 and FRK1 were monitored by RT-qPCR analyses. The mRNA levels are relative to the level of UBQ transcripts. Statistical significance of flg22 treatment on expression was assessed using a two-way ANOVA test and a Sidak’s multiple comparisons test. Asterisks indicate statistical significance (*: p<0.05, **: p<0.01, ***: p<0.001, ns: not significant). (E) IR-RMG1p/RLP43p lines exhibit increased Pto DC3000 titre. Five-week-old plants of Col-0, ros1-3, and two independent IR-RMG1p/RLP43p lines were dip-inoculated with Pto DC3000-GFP at 5 × 107 cfu ml−1. Bacterial titres were monitored at 3 days post-infection (dpi). Each data point represents bacterial titre extracted from a single leaf. Three leaves out of four plants per line and from three independent experiments were considered for the comparative analysis. Statistical significance was assessed using a one-way ANOVA test and Tukey’s multiple comparisons test.
-
Figure 6—source data 1
Original McrBC-qPCR, qRT-PCR, and bacterial propagation data for Figure 6B–E.
- https://cdn.elifesciences.org/articles/62994/elife-62994-fig6-data1-v2.xlsx
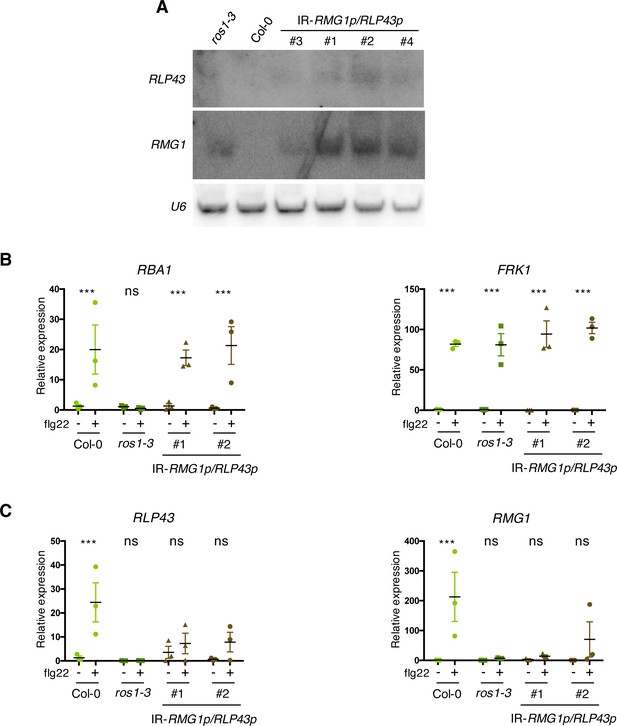
Artificial siRNA-directed remethylation of RMG1 and RLP43 promoters in the Col-0 background limits flg22-triggered induction of these genes.
(A) Accumulation of siRNAs at RLP43 and RMG1 promoters in IR-RMG1p/RLP43p lines was detected by low molecular weight northern blot analysis. U6 was used as a loading control. (B) Result from another independent experiment showing gene expression of RBA1 and FRK1 upon 6 hr of mock or flg22 treatments. RT-qPCR analyses of mRNAs from these genes were performed in Col-0, ros1-3, and the two IR-RMG1p/RLP43p independent transgenic lines treated with either mock (water) or 1 μM of flg22 for 6 hr. The mRNA levels are relative to the level of UBQ transcripts. Statistical significance of flg22 treatment on expression was assessed using a two-way ANOVA test and a Sidak’s multiple comparisons test. Asterisks indicate statistical significance (*: p<0.05, **: p<0.01, ***: p<0.001, ns: not significant). (C) Result from another independent experiment showing gene expression of RLP43 and RMG1 upon 6 hr of mock or flg22 treatments. RT-qPCR analyses of mRNAs from these genes were performed in Col-0, ros1-3, and the two IR-RMG1p/RLP43p independent transgenic lines treated with either mock (water) or 1 μM of flg22 for 6 hr. The mRNA levels are relative to the level of UBQ transcripts. Statistical significance of flg22 treatment on expression was assessed using a two-way ANOVA test and a Sidak’s multiple comparisons test. Asterisks indicate statistical significance (*: p<0.05, **: p<0.01, ***: p<0.001, ns: not significant).
-
Figure 6—figure supplement 1—source data 1
Original qRT-PCR data for Figure 6—figure supplement 1.
- https://cdn.elifesciences.org/articles/62994/elife-62994-fig6-figsupp1-data1-v2.xlsx
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Arabidopsis thaliana) | ROS1 | Arabidopsis.org | AT2G36490 | |
| Gene (Arabidopsis thaliana) | DCL2 | Arabidopsis.org | AT3G03300 | |
| Gene (Arabidopsis thaliana) | DCL3 | Arabidopsis.org | AT3G43920 | |
| Gene (Arabidopsis thaliana) | RMG1 | Arabidopsis.org | AT4G11170 | |
| Gene (Arabidopsis thaliana) | RLP43 | Arabidopsis.org | AT3G28890 | |
| Strain, strain background (Pseudomonas syringae pv. tomato) | Pto DC3000-GFP | Gift from Pr. Sheng Yang He (Duke University, US). | ||
| Genetic reagent (Arabidopsis thaliana) | ros1-3 | Penterman et al., 2007 | ||
| Genetic reagent (Arabidopsis thaliana) | ros1-4 | Nottingham Arabidopsis stock center (NASC) | N682295 | |
| Genetic reagent (Arabidopsis thaliana) | dcl2-1 dcl3-1 | Xie et al., 2004 | ||
| Genetic reagent (Arabidopsis thaliana) | ros1-3 dcl2-1 dcl3-1 | This study | ||
| Genetic reagent (Arabidopsis thaliana) | rmg1-1 | Nottingham Arabidopsis stock center (NASC) | N678063 | |
| Genetic reagent (Arabidopsis thaliana) | rmg1-2 | Nottingham Arabidopsis stock center (NASC) | N674117 | |
| Commercial assay or kit | MagneGST Pull-Down System | Promega | V8870 | |
| Commercial assay or kit | McrBC | New England Biolab (NEB) | M0272 |
Additional files
-
Supplementary file 1
DMRs position of 102 less-induced and 115 less-repressed ROS1 targets.
- https://cdn.elifesciences.org/articles/62994/elife-62994-supp1-v2.xlsx
-
Supplementary file 2
Primers used in this study.
- https://cdn.elifesciences.org/articles/62994/elife-62994-supp2-v2.jpg
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/62994/elife-62994-transrepform-v2.docx





