To promote reproducibility in scientific research, eLife encourages authors to include Research Resource Identifiers (RRIDs) within the materials and methods section of their papers. These persistent and unique identifiers help capture an unambiguous description of the research resources within articles.
We invited Anita Bandrowski, NIF Project Lead, UCSD and Founder & CEO, SciCrunch to contribute to Inside eLife and discuss why authors are choosing to include RRIDs in their papers and the impact this is having on reproducibility.
The Resource Identification Initiative (RII) is celebrating some major milestones: the total number of published papers containing RRIDs has surpassed 2,000; eLife and the Journal of Neuroscience have each published more than 250 papers with RRIDs, while other journals such as Cell and Neuron have published more than 100; and the number of published papers containing RRIDs is accelerating.
The number of journals publishing papers with RRIDs now tops 200, although these are mostly publishing RRIDs in small volumes and often without encouragement to their authors. At the other end of the spectrum are journals that are actively promoting RRIDs, including The Journal of Comparative Neurology, Cell, Neuron, Endocrinology, the Society for Neuroscience Journals, and eLife. eLife is a good example of what a complete implementation of the RRID guidelines looks like, so in this post I discuss how this happened and whether we can start to reveal some aspects of the RRID that in the future will be much more available as additional journals and authors follow the standard.
eLife started encouraging authors to include RRIDs in June 2016, stating the following: “To help promote the identification, discovery, and reuse of key research resources, we encourage you to include Research Resource Identifiers (RRIDs) within the Materials and methods section to identify the model organisms, cells lines, antibodies, and tools (such as software or databases) you have used (e.g. RRID:AB_2178887 for an antibody, RRID:MGI:3840442 for an organism, RRID:CVCL_1H60 for a cell line, and RRID:SCR_007358 for a tool).”
However, eLife’s instructions to authors are not strikingly different from other journals that encourage the use of RRIDs, so why do eLife authors appear to be so receptive to including them? One reason is that eLife was able to call upon some published examples from the archive, where early adopters had included them without the journal knowing: by adding the links to those papers, eLife had good examples to share with authors. Another reason is that eLife asks authors directly, during checks on papers at the point of resubmission, so when a paper is close to acceptance and the authors are already making other changes. Finally, during the production process, the eLife content processor searches the paper for “RRID” and then uses the SciCrunch API to convert the text into a URL. If this RRID does not resolve to a URL, this is queried during the proofing stage and can be corrected before publication.
Anindya Dutta from the University of Virginia, who published with eLife and included RRIDs ( http://dx.doi.org/10.7554/eLife.19084), said: “Besides helping other labs to reproduce our experiments, it will help subsequent members in my lab to get exactly what they need for their experiments." Judith Schmitz and Sebastian Ocklenburg (Ruhr University Bochum), from another eLife paper ( http://dx.doi.org/10.7554/eLife.22784), added: “We added RRIDs in our paper mainly for identifying software tools. Adding the identifier turned out to be very straightforward. We expect it to be a helpful resource for enhancing the transparency of not only our paper, but also countless other ones.”
Indeed, linking to biological resources is invaluable for scientists wanting to optimize a protocol or track down an issue in their own lab. The question of which exact fly or antibody is used in an experiment can mean days of grueling work at the bench, but this is saved if the reader can find the correct resource immediately. Even if a reader is not planning to repeat the experiment, knowing the exact resource could influence their interpretation of the results, for example if a cell line is known to be contaminated, or an antibody is known to be problematic, or a knockout is incomplete. The following screenshot below shows the RRID portal’s response to a query for a contaminated cell line, 1-5c-4:
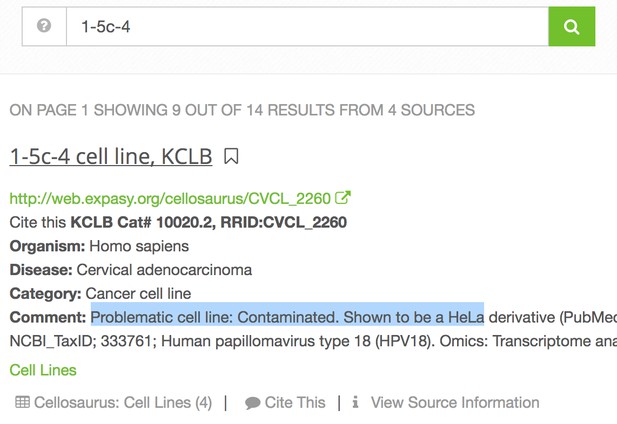
The International Cell Line Authentication Committee (ICLAC), chaired by Amanda Capes-Davis, was formed in 2012 to bring greater visibility to cell line misidentification and to promote awareness around authentication testing. The ICLAC’s Database of Cross-contaminated or Misidentified Cell Lines includes cell lines that are known to be cross-contaminated or misidentified. While not all vendors update the cell line contamination information regularly, RRIDs are synchronized by the Cellosaurus database, and as soon as the ICLAC releases new data, RRIDs are updated. The information is visible to authors using the cell line as a comment, alerting authors the cell line they used is contaminated.
At eLife, authors using cell lines must report their source, state whether the identity has been authenticated, state the authentication method (such as STR profiling), and report the mycoplasma contamination testing status. Authors are also asked to check the list of commonly misidentified cell lines maintained by the ICLAC before submission. Christopher Korch, a founding member of the ICLAC, thinks eLife authors “deserve sincere compliments for the efforts made to ascertain the identity of some of the cell lines they used by STR profiling. I am also glad to see that eLife is promoting cell line authentication by including the Cellosaurus/RRIDs CVCL numbers.”
The problems involved with publishing work with a contaminated cell line points to some interesting future directions and the real power of using RRIDs consistently across a broad swath of the literature. First, at least in eLife’s experience, it looks as though authors, when presented with information about contamination, are paying attention: they are using good cell lines, perhaps because they already were using good cell lines or perhaps because they paid attention to the note. Second, in the future, with many more journals using RRIDs, we should know almost effortlessly when there are reports of problems with reagents, which can have a serious impact on the interpretation of published papers.
Of course preventing the use of questionable reagents is not the only use of the RRID. We have the ability, especially as new journals join the initiative to ask simple questions efficiently. Which papers used this antibody in a western blot experiment, is currently an unreasonably difficult task, but with RRIDs implemented across many journals, the task would only involve putting in the RRID and the term western blot into PubMedCentral or google scholar, no matter what the antibody company choses to call itself this week and no matter whether the antibody has been discontinued or sold to another company.
Many journals are currently taking important steps to allow RRIDs, encourage the use of RRIDs or require the use of RRIDs and we welcome and encourage all of these steps in making science just a little more reproducible. To help journals and editors, the RRID initiative has gathered instructions to authors and put them up as samples for journals that would like to implement RRIDs, but are not sure how.
We welcome comments/questions from researchers as well as other journals. Contact staff [at] elifesciences [dot] org.




