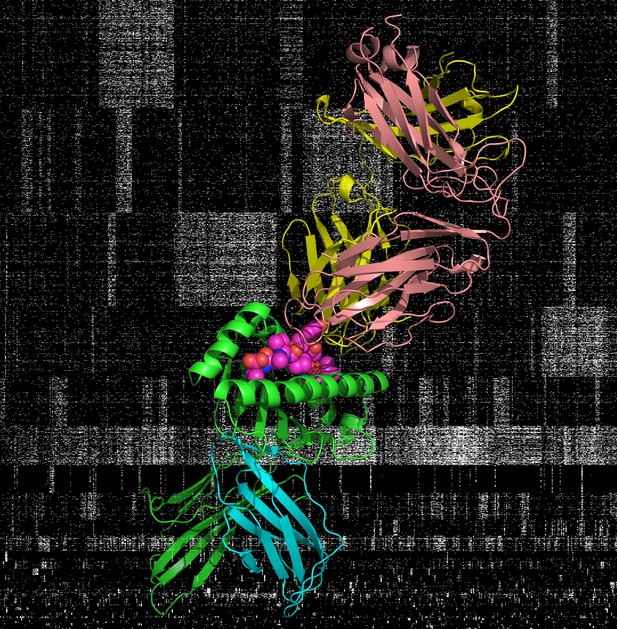
The crystal structure of a T cell receptor as it attaches to a protein fragment carried by a MHC. In the background is a matrix of T cell receptor occurrence patterns. Image credit: Phil Bradley (CC BY 4.0)
The immune system has two major ways of clearing up an infection. A rapid, first line of defense buys time while the second ‘adaptive’ response disposes of the threat with precision. The adaptive response takes longer to develop but once it has dealt with a disease, it remembers: the next time the body encounters the same threat, the immune system can respond much faster.
When cells are infected by a disease-causing microbe, like a bacterium or a virus, they start carrying fragments of that microbe on their surface. Immune cells known as T cells then recognize these fragments using proteins called T cell receptors. Each T cell has a different receptor, which is specific to a precise fragment of a particular microbe. After successfully clearing an infection, some of the T cells that were mobilized remain in the blood. These memory T cells, and their specific receptors, are a lasting trace of the infections a person has encountered in the past.
The exact portion of the microbial fragments that the T cells receptors can ‘see’ depends on another set of proteins, called MHC. These hold the fragments at the surface of the infected cells. The genes that code for MHCs are incredibly diverse, to the point that the exact combination of MHCs carried by a cell can be specific to an individual. However, different MHCs present different microbial fragments, and this changes which receptor can recognize the infection. At the level of a population, this mechanism makes it difficult to use T cell receptors to know exactly which diseases people had to face.
Here, DeWitt et al. look at the T cell receptor sequences of 666 healthy participants, as well as their MHC variants, to try to reconstruct their disease history. This revealed that many people have clusters of similar T cells receptors sequences that occur together; these could be linked to exposure to common viruses such as parvovirus, influenza, cytomegalovirus and Epstein-Barr virus. Furthermore, examining 3D structures of T cell receptors binding to fragments carried by MHCs helps to identify how changes in the sequence of the MHC can influence which receptor will be able to attach to the complex.
These results show that, despite the diversity and complexity of T cell receptors and MHCs, it is possible to spot patterns across people, and to start understanding how those patterns emerge. In addition to fighting body invaders, T cells can also use their receptors to recognize certain protein fragments carried by tumor cells. Improving our knowledge of T cell receptors and MHCs could give new insights to fight cancer.




