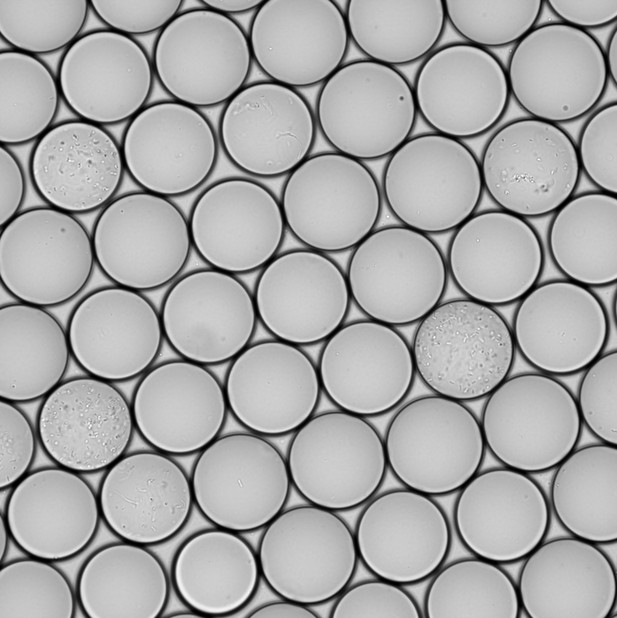
A new culturing system that isolates and grows gut bacteria in minuscule droplets of liquid. Image credit: Watterson et al. (CC BY 4.0)
The human gut is inhabited with hundreds of billions of bacterial cells from a wide range of families. This complex mixture of bacteria is part of the gut microbiome, along with other lifeforms such as viruses, archaea and fungi. As well as interacting with each other, the bacteria in the microbiome interact with our cells and available nutrients. Studying these interactions can help us understand how this community of bacteria influence health and disease.
One way to study the diversity of the microbiome is to take a sample, such as a section of stool, and perform DNA sequencing to determine which types of bacteria are present. This can reveal how the composition of the gut microbiome relates to our health, but cannot confirm whether these bacteria are the cause or the effect of most diseases.
To overcome this problem, researchers need to be able to grow pure strains of these bacteria in order to unravel their underlying mechanisms. For over a century, the conventional way to cultivate bacteria has been to grow them in a Petri dish. However, this method promotes the growth of more abundant, fast-growing bacterial strains. This results in a huge disconnect between the bacteria grown in a Petri dish and the diversity within the human gut, which is hindering our understanding of gut health and disease.
Now, Watterson et al. have built a machine that improves the speed and number of cultivated bacterial organisms, thus paving the way for more detailed investigations of the human gut microbiome. This new system works by growing bacteria in millions of miniscule droplets which can be physically separated to help the expansion of slower growing species.
Watterson et al. cultivated bacterial cells from a stool sample from a single donor using the droplet system and compared this to traditional culturing methods. The droplet technology increased the number of different organisms that were able to grow by up to four times, including those that were rare or slow-growing. Bacteria in the donor stool were then screened for populations that were resistant to antibiotics. This identified 21 antibiotic resistant bacteria which only grew in the droplets and not in Petri dishes.
This droplet-based technology will make it possible to study bacterial strains that were previously difficult to grow. Furthermore, this method could help identify whether stool from a donor contains any antibiotic resistant strains, which can lead to clinical complications once transplanted. In future, this new technology could be used in laboratories or hospitals to study the role of the gut microbiome in health and disease.




