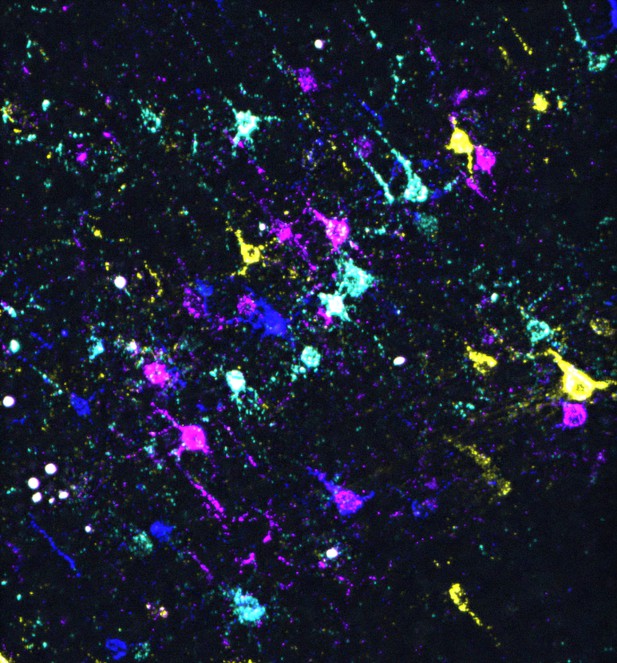
In situ sequencing of barcoded mouse neurons in a brain slice. Image credit: Zhang, Jin et al. (CC BY 4.0)
In the brain, messages are relayed from one cell to the next through intricate networks of axons and dendrites that physically interact at junctions known as synapses. Mapping out this synaptic connectivity – that is, exactly which neurons are connected via synapses – remains a major challenge.
Monosynaptic tracing is a powerful approach that allows neuroscientists to explore neural networks by harnessing viruses which spread between neurons via synapses, in particular the rabies virus. This pathogen travels exclusively from ‘postsynaptic’ to ‘presynaptic’ neurons – from the cell that receives a message at a synapse, back to the one that sends it. A modified variant of the rabies virus can therefore be used to reveal the presynaptic cells connecting to a population of neurons in which it has been originally introduced. However, this method does not allow scientists to identify the exact postsynaptic neuron that each presynaptic cell is connected to.
One way to bypass this issue is to combine monosynaptic tracing with RNA barcoding to create distinct versions of the modified rabies virus, which are then introduced into separate populations of neurons. Tracking the spread of each version allows neuroscientists to spot exactly which presynaptic cells signal to each postsynaptic neuron. So far, this approach has been used to examine synaptic connectivity in neurons grown in the laboratory, but it remains difficult to apply it to neurons in the brain.
In response, Zhang, Jin et al. aimed to demonstrate how monosynaptic tracing that relies on barcoded rabies viruses could be used to dissect neural networks in the mouse brain. First, they confirmed that it was possible to accurately detect which version of the virus had spread to presynaptic neurons using both in situ and single-cell RNA sequencing. Next, they described how this information could be analysed to build models of potential neural networks, and what type of additional experiments are required for this work. Finally, they used the approach to identify neurons that tend to connect to the same postsynaptic cells and then investigated what these have in common, showing how the technique enables a finer understanding of neural circuits.
Overall, the work by Zhang, Jin et al. provides a comprehensive review of the requirements and limitations associated with monosynaptic tracing experiments based on barcoded rabies viruses, as well as how the approach could be optimized in the future. This information will be of broad interest to scientists interested in mapping neural networks in the brain.




