We are delighted to showcase three projects entered for the eLife prize at Hack Cambridge Recurse: eXplore, Knowledge Direct and SciChat. The winner of the eLife prize was eXplore. These hackathon projects are still early stage, and each team welcomes contributions and feedback via the respective GitHub repositories.
eXplore
Team members:
- Charlotte Guzzo – PhD student in Biological Sciences, Sanger Institute, University of Cambridge
- Will Jones – PhD student in Mathematical Genomics and Medicine, University of Cambridge
- Patrick Short – PhD student in Mathematical Genomics and Medicine, University of Cambridge
Most genetic data holders do not have a background in genetics and do not understand scientific papers. They often wish to know how research relates to them specifically and what the key takeaways are. Charlotte, Will and Patrick were interested in enabling the non-expert reader to fully appreciate their genomic data by accessing content that is relevant to them and easily understandable, without being restricted to information provided by third-party data providers. According to team eXplore, this is particularly important in engaging public opinion and encouraging participation in research.
At Hack Cambridge, the eXplore team aimed to offer a reading experience that is tailored to each individual user’s genotype and explains the key points and limitations in plain language. Using the eLife Lens API, they designed a tool that personalises the reader’s experience when exploring scientific information. To demonstrate proof of concept, the team wrote a simple article on how genetics impacts taste preferences. Using a team member’s own genetic data from 23andMe, they delivered personalised information about a gene variant when it was mentioned in the text.
A video demonstration of eXplore. Source: https://youtu.be/RKUurAF5sdI.
As well as developing more curated reports to help users understand the literature, the team would like to empower users further by helping them to navigate the references themselves and critically review the literature. To do that, they are planning to work on a Chrome extension that would highlight key points in a scientific paper, and mark the parts directly related to the user’s genotype. Using the extension, users would be able to explore research articles cited in the curated reports and immediately find the content that is relevant to their genotype.
The eXplore team welcomes contributions from scientists willing to develop reports and from developers willing to help with developing the pipeline and data integration. You can explore the roadmap and get in touch with the team via their GitHub page.
Reflecting on their experience, the eXplore team noted:
"The eLife prize was very aligned with what drives us as scientists. Making science communication open to all is something we had been discussing for a while, and we felt strongly about working on a project that would make scientific research more approachable and relevant to everyone. The most valuable part of the weekend was to work together in such a focused way and to meet brilliant participants from all over the world."
Resources used for the hack:
The eLife Lens API was used to display the prototype; the 23andMe API was used to pull data from 23andMe users; and the PubMed Central API was used to retrieve scientific papers containing specific single nucleotide polymorphisms for the curated reports. The team used Python/node.js to write the program.
The source code is available at https://github.com/pjshort/eXplore/.
Knowledge Direct
Team members:
- Jack Hughes – Undergraduate in Computer Science, University of Birmingham
- Jeremy Minton – PhD student in Mathematics, University of Cambridge
- Veronika Siska – PhD student in Computational Biology, University of Cambridge
- Edward Stevinson – MSc student in Artificial Intelligence, University of Edinburgh
Entering a new academic field, or researching beyond your comfort zone, can be challenging. Existing literature search tools present the most relevant publications, but without any guidance as to the background or context required to understand the material or its significance in full. Further, constructing efficient and targeted searches using keywords requires a degree of familiarity with the field already. The researchers in the Knowledge Direct team have previously found conducting a literature review to be time consuming, painful and frustrating. Beyond the inconvenience, they think the traditional literature discovery process is inefficient, limits deeper collaborations and prevents amateur contributions in research.
At Hack Cambridge, the team wanted to help researchers become familiar with new fields much quicker by collecting metrics to construct an efficient path through the literature. To this end, they developed Knowledge Direct, a literature search engine to get you from what you know, to what you want to know, as efficiently as possible. This web application produces a network of publications, identifies your current knowledge, and presents a path through the literature to help you become familiar with a new field. It helps the user to understand a collaborator’s publication or check key publications for a literature review quickly and efficiently.
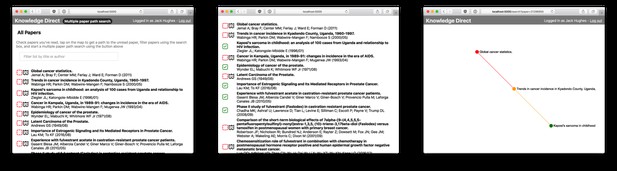
The Knowledge Direct interface (demo). The user searches for and selects papers that they have already read. Next, they click on the map next to an article from a new field they would like to explore. The result is the most efficient pathway through the literature that builds familiarity from what the user already understands to the desired endpoint article.
For the Knowledge Direct team, Hack Cambridge Recurse was an opportunity to learn about a selection of machine-learning techniques and produce a web application. When asked what led her to attend the hackathon, Veronika said:
“Most importantly, I just really love overnight coding: it's intense and engaging. I also got to work on the initial, most exciting phase of a completely new project, which is a rare occasion during a PhD. Finally, I was hoping to learn something new – in this case, handling publication data and applying techniques from network science to a real problem.”
For the future, the application could be taken forward as either a digital service or an open-source project. In order to achieve either of these, developing the underlying network construction would be essential. The Knowledge Direct team encourages interested contributors to get in touch via the GitHub repository.
Resources used for the hack:
The PubMed Central API was used to access a large body of academic literature.
The source code is available at https://github.com/knowledge-direct/knowledge-direct.
SciChat
Team members:
- Nils Eling – PhD in Molecular Biology, University of Cambridge, Cancer Research UK
- Raghd Rostom – PhD student in Molecular Biology, Wellcome Trust Sanger Institute, University of Cambridge
- Dimitrios Vitsios – PhD student in Bioinformatics, University of Cambridge, European Bioinformatics Institute (EMBL-EBI)
- Omar Wagih – PhD student in Computational Biology, University of Cambridge, European Bioinformatics Institute (EMBL-EBI)
The complex nature of modern scientific findings makes them difficult to communicate beyond an expert, scientific audience. In addition, opportunities for the public to interact with experts are limited. Nil, Omar, Raghd and Dimitrios believe that everyone should benefit from the outcome of scientific studies.
At Hack Cambridge, the team developed SciChat to bridge the often large gap between the general public and scientific research. SciChat is a live-chat tool that connects members of the public with scientists in areas of interest using simple tags. Members of the general public can very easily and anonymously be paired with corresponding scientists through a live one-to-one chat where they can discuss the topic of interest in an informal, conversational manner. The platform also helps scientists learn how to communicate their findings in an understandable fashion.
A video demonstration of SciChat. Source: https://www.youtube.com/watch?v=PdNuhiDcmUM.
The SciChat team is hoping to refine the current website by improving usability and integrating additional features. In addition to connecting scientists with non-scientists, they hope to allow researchers from different fields to communicate with one another. Scientists would be able to display a profile including information about their research, along with links to publications and professional websites. The team would also like to add a reputation system, allowing academics to rate the engagement of the user they chatted with, and vice versa. Such data would allow the community to identify the most committed users, and would enable more sophisticated pairing. In addition to this, there could be potential to highlight trending topics and create user statistics, such as average rating and number of chats. The SciChat team welcomes contributors and can be contacted via the GitHub repository. They commented:
“Despite working in the same field, all of us come from a variety of backgrounds ranging from cancer biology to computer engineering. The hackathon allowed us to engage in interdisciplinary thinking, while exposing ourselves to frameworks and APIs that were new to us. We believe that hackathons in general are one of the best environments to meet with brilliant people, collaborate within a group to achieve a goal in a limited timeframe, and to learn new technologies.”
Resources used for the hack:
The Google Firebase cloud database was used as a backend for SciChat, and peer-to-peer browser technology, PeerJS, was used to power the chat engine. The logo and vector graphics used were modified from freepik, and the user interface was custom-built based on Bootstrap.
The source code is available at https://github.com/omarwagih/scichat.




