We are pleased to bring attention to a new and exciting open-source project called Binder, created by scientists at HHMI’s Janelia Research Campus.
We are looking for new ways to exploit the flexibility of digital media, to present and make available all kinds of data and analysis, encouraging greater transparency in research and enabling the scientific community to build upon published results more effectively.
Binder is a website and collection of open-source tools for building and executing reproducible computational environments. Binder makes it easy to include an interactive version of your analysis, with the supporting data and code, alongside a published paper. An example has recently been included with a neuroscience research article by Sofroniew et al. 1
Blogpost by Jeremy Freeman and Andrew Osheroff, developers of Binder, at HHMI’s Janelia Research Campus
Modern science depends on data analysis. From neuroscience to genomics, to cancer research, scientific conclusions are often several stages removed from raw data, and reflect extensive data processing and statistical analyses.
Yet in the traditional academic paper, we can only show a small sample of raw data, and report just a few of many possible summary statistics. We have to describe our analyses in compact paragraphs of plain text sprinkled with equations — an opaque starting point when trying to reproduce an analysis. Data and code, if shared at all, are appended to the paper as an afterthought, without ensuring that they are easy to reuse.
Why does this matter? Scientific progress depends on replicating and validating the work of others. And replicating what someone else has done is often the starting point for scientific collaboration.
Several open-source tools can be used to help address the challenges of sharing and reproducing scientific analyses. The Jupyter notebook is a coding environment that runs in a web browser and lets users create computational “narrative documents” that combine code, data, figures, and text in a single interactive, executable document. These notebooks are easy to write, support many programming languages, and are already being used in science, journalism, and education.
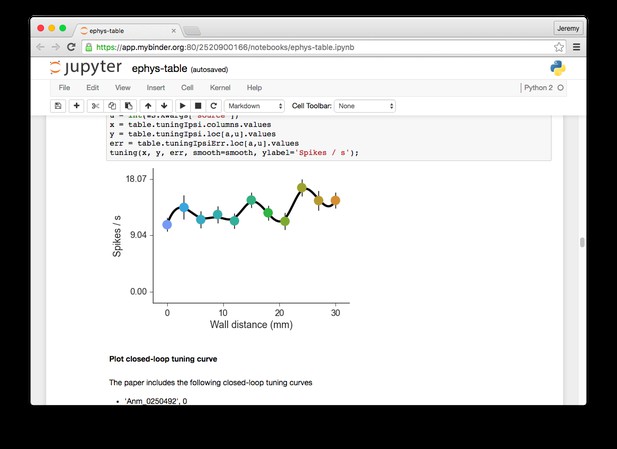
An example executable notebook using Binder, from Sofroniew et al. 1.
GitHub is a website for collaborative code development, built on top of the version-control system git. GitHub makes it easy to track changes to code over time, especially when multiple contributors are working on the same project. Putting data, code, and notebooks into a GitHub “repository” is a terrific way to share and organize scientific analyses.
The GitHub repository for Sofroniew et al.'s project 1.
But just providing our code, data, and notebooks alongside a paper isn’t enough — what ran on my machine might not run on yours. We can share our computer configurations, but setting up a new machine the exact same way can be challenging and unreliable.
We designed Binder to make it as easy as possible to go straight from a paper to an interactive version of an analysis.
How it works
To use Binder, you only have to put the code, data, and Jupyter notebooks that you are already using for analysis on your machine into a GitHub repository, and provide Binder with the link.
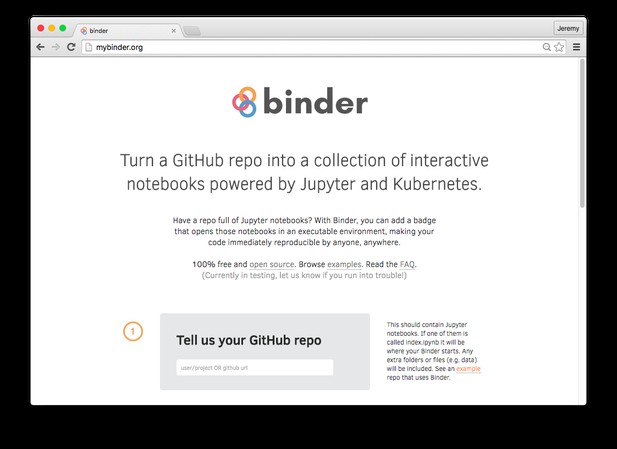
To create a Binder, enter your GitHub repository URL at mybinder.org.
Binder will then build an executable environment that contains all the necessary dependencies to run your code, and can be launched by clicking a link in your web browser. Now, with one click, anyone can immediately inspect the raw data, recompute a statistic, regenerate a figure, and perform arbitrary interactive analyses.
To make this work, Binder uses existing, robust tools wherever possible. Along with Jupyter and GitHub, Binder leverages two open-source projects under the hood to manage computational environments — Docker builds the environments from a project’s dependencies, and Kubernetes schedules resources for these environments across a cloud compute cluster.
A key use case for Binder is sharing analyses alongside traditional journal publications, and a few great examples of that already exist. A recent paper on neural coding in the somatosensory cortex by Sofroniew et al. 1 in eLife used Binder to share data and analyses of neural recordings. Another example is a recent paper in Nature by Li et al. 2 on robustness to perturbation in neural circuits, which used Binder to share simulation results from a computational model. Notebooks demonstrating the discovery of gravitational waves from the LIGO group were turned into a Binder, which has been by far our most popular example.
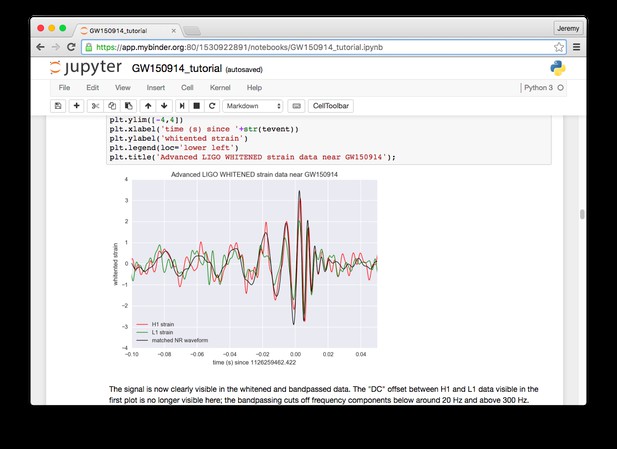
A Binder for exploring gravitational waves data from the LIGO group.
We’ve also seen Binder used in domains we didn’t expect. Outside of science publications, Binder has been used to make analyses for news stories more reproducible, and even to make an entire book on data science executable. It’s also become a popular way to run courses or tutorial sessions, because students can launch tutorials straight from their web browsers, without wasting precious time configuring dependencies. Physicists at CERN use it to showcase demos of their ROOT analysis framework.
Next steps
With more than 1200 reproducible environments already built by its users, Binder has proven useful and we’re excited by its potential — but much work remains, both on Binder itself and the underlying technologies, especially for making it better suited to scientific publishing.
Here are a couple of directions we’re excited about:
We currently maintain a public Binder deployment, hosted by our lab at HHMI Janelia Research Campus and running on Google Compute Engine, designed for open source and open science. But we’ve recently made it easy for others to deploy custom versions of Binder on their own compute infrastructure. We hope this can provide a way for publishers to deploy and host Binders with guaranteed availability for their readers.
We currently recommend users put data in their GitHub repositories, but git was designed to keep track of code, not data, which often consists of heterogenous files in a variety of formats, rather than plain text. We are collaborating with the team behind a new peer-to-peer data sharing and versioning project called Dat, and hope to integrate it with Binder.
We want to work on new ways to integrate static papers and interactive notebooks. Even with Binder, these remain separate and very different kinds of documents. Ideally, we would have unified interfaces that give readers the option to engage with code and figures at whatever level of detail they desire.
Thanks to Fernando Perez, Brian Granger, Kyle Kelley, Min RK, and Max Ogden for help, inspiration, discussion, and ideas.
References:
- Sofroniew, N. J., Vlasov, Y. A., Hires, S. A., Freeman, J. & Svoboda, K. Neural coding in barrel cortex during whisker-guided locomotion. eLife 2015;4:e12559. http://dx.doi.org/10.7554/eLife.12559
- Li, N., Daie, K., Svoboda, K. & Druckmann, S. Robust neuronal dynamics in premotor cortex during motor planning. Nature 532, 459-464 (2016). http://dx.doi.org/10.1038/nature17643




