By Naomi Penfold, Innovation Officer, and Maria Guerreiro, Journal Development Editor, eLife.
On July 11, we are joining researchers, publishers, repository vendors, policymakers and research data managers for a workshop to discuss use cases and workflows for the deposition and preservation of research software. The workshop, supported by Jisc, has been organised by the Software Sustainability Institute and follows a planning workshop and report earlier this year summarising the existing options for research software deposition and archiving.
At the workshop, we are presenting eLife’s perspective as a publisher in the life sciences, following preliminary work to understand current behaviour by researchers in this regard, alongside our requirements and those of the research community as a whole. A summary of our perspective follows underneath the slides for the presentation:

The slides are available on Google Slides and Figshare with DOI: 10.6084/m9.figshare.6799097.
Software and code form an integral part of research across the broad spectrum of the sciences, including, to a growing degree, biological research. The majority of authors share code with eLife through links to online version-controlled repositories: while this is almost exclusively through GitHub, sites such as GitLab, Assembla and Bitbucket have also been used. Some authors have deposited their code in an external data repository, such as Dryad, when analysis code accompanies a large dataset (for example, see http://datadryad.org/resource/doi:10.5061/dryad.854j2). The dominance of GitHub is due to a vast majority of eLife authors already sharing code there prior to making their revised submission, which is the stage in the process when eLife staff check for software availability. Those who cite software not already available are prompted by our editorial team to deposit the code, and authors will usually either use GitHub or upload the files to the eLife submission system. For authors who have software deposited on their lab web pages, we ask them to deposit it elsewhere too, ideally using version control.
We also receive source files (Python scripts, for example) directly to our submission system. In this case, the files are stored as zip folders in our content delivery network (CDN; through Amazon Web Services) and they are made available to download via links from the article on our HTML publishing platform. The most frequent languages used for research presented in eLife are MATLAB, Python, R and C++, and we also receive Jupyter Notebook files and subject-specific formats such as PyMOL files.
As a publisher, it is our responsibility to disseminate research outputs, including code and software, as an integral part of the publication process and to preserve these assets for future reading and reuse wherever possible. Since February 2017, it has been our policy to fork research software stored in online version-controlled repositories to our GitHub account upon acceptance, providing a snapshot to accompany the publication in case of onward development or deletion. Since the introduction of the policy, we have forked nearly 300 repositories. The eLife forked copy is shared as an in-text citation following the citation of the authors’ original source repository, the latter of which is included as a formal reference too.

The current workflow has a number of benefits. Forking to our own GitHub repository represents the route of least resistance for the majority of researchers writing, sharing and reusing research software: it’s based around GitHub. Because we are involved in the process, we can use our leverage as a publisher to encourage best practises, including not only asking authors to share code in the first place, but also to do so with permissive licensing and to add formal software citations to their article. This complements our broader efforts to encourage the sharing of research data and materials whenever possible to facilitate greater reproducibility in science: we have introduced the eLife Transparent Reporting Form and the Key Resources Table in recent years as well. We also prefer to reduce the burden on the author, and therefore take responsibility for forking and adding the inline citation for this to the manuscript. Alternative author-driven approaches do not provide this opportunity to influence, and they also leave the permanence of the archive copy in the hands of the author instead of the publisher.
However, we are also aware of limitations. For one, the process is reliant on GitHub, which is a centralised and privately owned service, meaning our process is dependent on GitHub remaining a free service for open content. While we expect this to remain the case, it is sensible to assess community-owned alternatives and explore sustainability options for those.
In addition, our current process does not cater for source code files shared directly with the journal: these remain stored in the journal’s CDN and are not archived externally. We are interested to understand how the community perceives the advantages and disadvantages of research code being shared via supplementary files, in contrast with that shared through online repositories associated with version-controlled systems, such as GitHub.
Further, this process requires manual efforts within our editorial and production workflows, with our staff estimated to spend more than two hours a week on it, depending on the number of publications in process at the time. Email correspondence with the authors contributes roughly 25% of this time cost.
Processes that require staff time may not be sustainable as research practises develop and become more common. We are interested in developing workflows and processes that would reduce this burden for all publishers and so improve the cost-efficiency of publishing, while not impacting on adherence to responsible research and publishing practises. We certainly expect there are parts of our current process that could be automated and are interested in alternative processes that provide a similar or better solution, with less publishing staff and author burden.
To consider our requirements for any process development, we have investigated our base requirements as a publisher alongside guidance from the research community. Earlier this year, we ran an initial poll using social media to get an impression of researcher requirements for preserving research software.
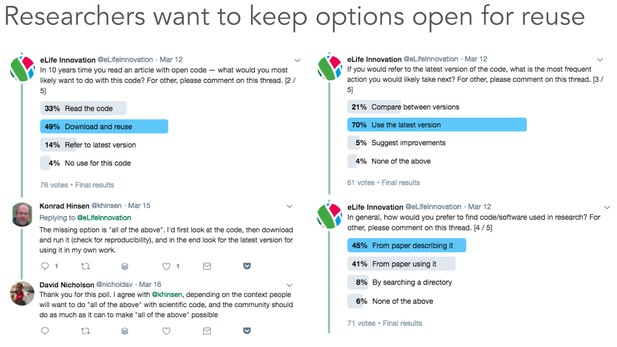
Twitter poll conducted from March 12 to March 19 inclusive, from @eLifeInnovation.
We received over 70 responses and learned that researchers would like flexibility over how they would be able to interact with archived research code in the future: they wish to be able to read, download and reuse 10-year-old research code. Only 4% of 76 respondents reported having no use for this code. At the moment, researchers predominantly access these assets directly from the academic literature rather than through centralised search services. These results suggest publishers and repository vendors must continue aiming to preserve research software in a way that does not restrict how it could be used in years to come.
In addition, and as demonstrated by existing researcher practises, any software deposition and preservation process will need to cater for a diverse range of sources and formats of research software. We welcome discussions about the preservation of more complex research software-related assets, for example pipelines that combine multiple scripts and require contextual information, and newer file formats that combine code with data and text, such as the dockerised reproducible research bundles shared with CodeOcean and the reproducible document archive format (Dar) being developed by Substance. Complementary to the suggestions made in the previous workshop and report on this matter, we wish to work with the community to facilitate the preservation of all assets required to execute research code in the future, such as ensuring clear documentation of dependencies and the sharing, deposition and preservation of container files such as Dockerfiles.
Finally, we include the following requirements from a publisher’s perspective:
- As mentioned above, we prefer to minimise additional work for authors and journal staff.
- We think it is important that accurate metadata is collected and maintained at source and are committed to complying with the software citation draft principles (Smith et al., 2016) agreed by the FORCE11 software citation working group, such as by including sufficient information in software citations: title, authors, location, URL and version/commit number.
- Any archived assets must persist and remain retrievable from the archive location for as long as we have the technology to access the files.

Five key requirements for software preservation by eLife. Image credits, all CC-BY from the Noun Project unless otherwise stated (left to right, top to bottom): Work by BGBOXXX Design; Scale by Amelia; Reuse by Desbenoit, FR (Public domain, CC0); tags by Thomas Helbig; archive by Yuri Mazursky; Download by Landan Lloyd.
We believe the process of research software preservation presents an opportunity for the community to leverage the publishing process to encourage responsible behaviours in science, including transparency and reproducibility. From our investigations, we have identified a number of requirements for such a process and can see both benefits and shortcomings of eLife’s current workflow. Outstanding questions include where the responsibility lies for the review of software that research depends upon and whether the dominance of GitHub as a platform for documenting and sharing research software presents a risk for the research community. As part of the discussions, we hope to understand how the community would evaluate the value add of any further process development measured against the potential cost and effort.
eLife is open to testing new processes that meet our requirements as a publisher, and those of the research community, and we welcome ongoing discussions about this with interested parties.
We thank Mike Jackson, Neil Chue Hong and the teams at the Software Sustainability Institute and Jisc for the opportunity to contribute to these discussions, as well as all members of the research community who have contributed to our investigations and process development so far.
References:
Smith AM, Katz DS, Niemeyer KE, FORCE11 Software Citation Working Group. 2016. Software Citation Principles. PeerJ Computer Science 2:e86. DOI: 10.7717/peerj-cs.86.
We welcome comments, questions and feedback. Please annotate publicly on the article or contact us at editorial [at] elifesciences [dot] org.
For the latest in published research plus papers available in PDF shortly after acceptance sign up for our weekly email alerts. You can also follow @eLife on Twitter.




