By David Ciudad, Data Scientist
Citation records have already been used to interrogate citation behaviour (such as Greenberg, 2009), enrich the information provided with a citation (see also Di Iorio et al., 2018, for current work considering how to present this information to the reader) and explore a researcher’s scholarly network, for example. With the amount of open citation data growing, driven by the Initiative for Open Citations (I4OC) and OpenCitations with the Open Citations Corpus, we are interested in how these data could be used to derive a deeper understanding of the impact of a research paper. We have also been experimenting with the application of data science to open science, to create openly available insight and tools that add value. And so we have embarked on a data science project to investigate how natural language processing (NLP) techniques could provide insight into the context of scientific citations available as open citation data. In particular, we are asking whether data science techniques could reveal where a citation occurs within the paper’s narrative and whether the citation has a positive, negative or neutral sentiment (citation polarity). This work began as a short project through the ASI data science fellowship programme. Here, I report back from this project.
Data sources and methods
The aim of the ASI project was to develop a pipeline for understanding how a paper was cited – in which section and with what statement. Retrieving this information required access to the full-text record of the paper in which the citation was made (the citing paper). As this was a pilot investigation, I focused my analyses on highly cited papers: this gave me the best chance of finding cited papers with a sufficiently large available sample of full-text records for their citing papers from which to derive useful conclusions. Daniel Ecer, eLife Data Scientist, had previously compiled citation counts from Crossref’s open citation data – I focused on the papers with citation counts greater than 500 there, as well as identifying particularly highly cited papers using Google Scholar, since not all citation records are open.
To retrieve the full-text records of the citing papers, I used the PubMed Central Open Access Subset (PMC OAS), a collection of open-access scientific papers – currently including approximately 1.7 million papers – that can be accessed through an FTP service. I searched the PMC OAS for mentions of the cited paper DOI (as it should appear in the reference section) and excluded any self-hits (the cited paper itself). Not all citing papers are open access: the number of full-text records of citing papers available for cited papers was highly variable; some had none, some had a good proportion. The publication date of the cited paper was a factor, since open-access publishing only began in earnest in recent decades. I decided to only consider cited papers with 10 or more open-access full-text records that cite them to ensure I had sufficient information to work from.
For highly cited papers identified from open citation data and Google Scholar, open-access full-text records of citing papers were retrieved from PubMed Central and used in citation context analyses.
Once I had a dataset of cited papers and citing paper full texts, I could begin to ask questions of the data, including:
- Does the location of a citation within a citing paper provide context as to why a paper has been cited?
- Can NLP be used to indicate whether a citation has a positive, negative or neutral sentiment?
The Jupyter notebooks used for this work are openly available at https://github.com/elifesciences/citation-context. To run the analyses described below yourself using a DOI or list of DOIs of your choosing, follow https://github.com/elifesciences/citation-context/blob/master/front.ipynb. Below, I demonstrate some observations found using this pipeline.
In which section has a paper been cited?
The first contextual information we explored for the citation was where in the citing paper the citation was included – in the introduction, methods, results or discussion. We hypothesised that the section in which a citation appears could be informative: do methodological advances get cited more in methods sections of subsequent papers? Are important findings cited more in the introduction or discussion as work that is being built on? Evidence to support these hypotheses has recently been reported for a corpus of works in the Library and Information Sciences: Taskin and Al, 2018, found that the sentiment polarity (positive, negative or neutral) and citation purpose (to acknowledge a method used, validate previous work, define a concept and so on) was related to the section in which the citation appears. For example, both this study and investigation of the PLoS corpus (Bertin et al., 2016) find that negative-sentiment citations are predominantly found in the introduction, results and discussion, and not in the methods section.
In order to ask these questions for the corpus collected for this project, I extracted the location of a citation within full-text records. First, I developed a program to identify the different sections of papers available in the PMC OAS papers. The program looks for different sections – introduction, conclusions, discussion, references – by searching for the matching tag in the XML or by looking for appropriate section headings in the text. The methods, materials and results sections are labelled less consistently across journals, so here I group them as ‘main text’. The main text is the output after the XML front section, abstract and other identified sections are removed. A similar extraction has been performed across the PLoS corpus (Bertin et al., 2016); the program presented and shared here is agnostic of journal JATS-XML flavour.
With the program, I was able to identify the following sections (the example sentences below were obtained by analyzing Rountree et al., 2012):
| Section name | Example XML found |
|---|---|
| Introduction | <sec><title>Introduction</title><p>Life expectancy in people with Alzheimer's disease (AD) is, overall, shorter than what is expected in age-matched, cognitively normal seniors and may be influenced by age, disease severity, general debility, extrapyramidal signs, gender, and race or ethnicity [<xref ref-type="bibr" rid="B1">1</xref>-<xref ref-type="bibr" rid="B4">4</xref>]. |
| Main text | <sec sec-type="materials|methods"><title>Materials and methods</title><sec><title>Participants</title><p>Informed consent was received from all patients involved in the study. |
| Discussion | <sec sec-type="discussion"><title>Discussion</title><p>The median survival time of this cohort with probable AD diagnosis was 11.3 years from the onset of symptoms. |
| Conclusions | <sec sec-type="conclusions"><title>Conclusions</title><p>In this large AD cohort, survival is influenced by age, sex, and a calculable intrinsic rate of decline. Disease severity at baseline, vascular risk factors, and years of education did not influence time to death. |
Using this program, I could analyse in which section(s) and how often a paper was cited within papers available from the PMC OAS. Here are the results for two highly cited papers:
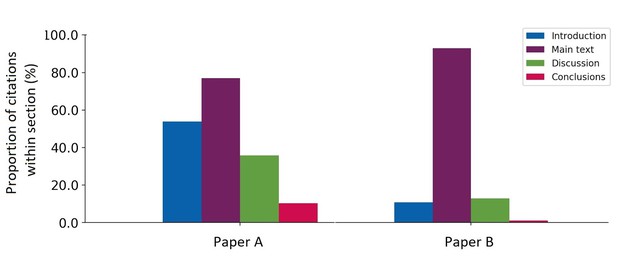
Citation fingerprint of two highly cited papers. For paper A, 39 citing papers were analysed (6% of total citation count 631, according to Google Scholar); for paper B, 94 citing papers were analysed (3% of total citation count 3,129, according to Crossref’s open citation data).
This analysis provides a fingerprint for the cited paper that may be informative for the context of the citation. For instance, references to paper A appear within most sections, but most frequently in the introduction and ‘main text’, whereas paper B has mainly been cited in the ‘main text’. Given the ‘main text’ includes the methods section, this may be an indication that paper B describes a new method or technique that has then been applied in future work, whilst paper A describes a result or research question that has been built on in subsequent work. In order to interrogate this assumption, I sought to understand the work being cited and the language used to cite it.
Extracting the citing sentences
For highly cited papers A and B, I retrieved the citing sentences from the full texts available from the PMC OAS. From all these sentences, I identified the most representative citing sentence within each paper section: this was the sentence containing the greatest number of ‘most frequent’ words used in all citing sentences for that paper in that section. Where there were multiple sentences meeting this criteria, the shortest one was selected. The outcome was a list of sentences (one per section) that could be used to understand the context of the citation at a glance.
For example, for paper A:
| Section | Proportion of citations within section (%) | Representative sentence |
|---|---|---|
| Introduction | 51 | “Reports of a newly-discovered gammaretrovirus (xenotropic murine leukemia virus-related virus: XMRV) in patients diagnosed with prostate cancer and chronic fatigue syndrome (CFS) have attracted the attention of investigators throughout the retroviral research community” (Smith, 2010) |
| Main text | 73 | “Thus there was no linkage between viral DNA detection and RNASEL R462Q in our clinical samples” (Sakuma et al., 2011) |
| Discussion | 34 | “Our results contrast with the high rate of XMRV detection reported by Lombard et al. among both CFS patients and controls, but are in agreement with recent data reported in two large studies in the UK and in the Netherlands” (Switzer et al., 2010) |
| Conclusions | 10 | “XMRV, originally identified in RNase L-deficient patients with familial prostate cancer, has gained interest since recent work showed its protein expression in as many as 23% of prostate cancer cases and XMRV-specific sequences were detected in PBMCs of 67% patients with chronic fatigue syndrome” (Fischer et al., 2010) |
Note that the sum of the proportions is greater than 100% since some papers will be cited more than once per citing article and in more than one section, and the citation count does not include these repetitions.
A quick reading of these sentences begins to suggest why and how this paper has been cited: paper A seems to be a report of a new virus (XMRV) in patients with chronic fatigue syndrome (CFS) and some citing papers appear to refute the link between this virus and CFS. In fact, paper A is Lombardi et al., 2009. This paper reports the detection of an infectious retrovirus (XMRV) in blood cells of patients with chronic fatigue syndrome (CFS) and suggests that XMRV can be a contributing factor in the pathogenesis of CFS. This paper was retracted because the results were not reproducible. Perhaps highlighting negative sentiment in citations may serve to provide early indication of concerns in the scientific literature. This is an angle we are now exploring.
Paper B is Lawton and Brody, 1969. This paper tests different scales used to assess functioning of older people and proposes standardised scales to be used in future experiments. Its representative citing sentences are as follows:
øtableøSection Proportion of citations within section (%) Representative sentence
This inspection is still a manual process, but it could be possible to automate the distinction between methodological mentions (as for paper B) and scientific discourse (as for paper A) by training an algorithm on pre-classified data. Further, it may be possible to use sentiment analysis to automate the detection of negative language (i.e. a scientific dispute, as for paper A).
Sentiment analysis for science
There have been several efforts to understand the sentiment of citations based on manual classification (for example: Athar, 2011; Athar, 2014; Xu et al., 2015; and Grabitz et al., 2017). An automated approach would increase the scale at which citations could be classified and broaden the application of this information. The idea of using artificial intelligence to automatically classify the context of a citation is over half a century old (Garfield, 1965). Several groups have now presented supervised machine learning approaches to classify citations based on extensive schema that go beyond identifying the basic polarity of sentiment: positive, neutral and negative (Teufel et al., 2006; Angrosh et al., 2014). It has been suggested that comprehensive classification based on schema or ontologies is not necessary for some applications of semantic tagging of citations: for example, understanding citation bias in the biomedical literature only requires an understanding of the polarity of the citation sentiment (Yu, 2013).
Whilst automated sentiment analysis tools are now available in the data scientist’s toolkit, they have been designed to analyse the highly emotive language used on social media. When applied to scientific texts, I found that existing sentiment analysis tools were insufficient to identify positive, neutral and negative language: the language used in science is more formal and less emotive than that used on social media. In scientific literature, neutral sentiment dominates and negative citations are even rarer than positive ones (Taskin and Al, 2018). Further, in a recent attempt to classify citations in the Turkish information sciences domain, the authors found that the accuracy of a Naive Bayes algorithm was diminished when neutral sentences were included (Taskin and Al, 2018). The authors called for an improved sentiment dictionary for scientific works to improve the accuracy of the algorithms. Therefore, we are interested in developing a new corpus of words or word structures (such as pairs of words) used for criticising, refuting or supporting comments in science, so that we can adapt sentiment analysis tools to identify positive, negative and neutral language used in scientific discussion in a more automated fashion than manual classification.
Much of the work conducted thus far has relied on manual annotation of citations in the test set. We are interested in building a fully automated sentiment dictionary for science. As indicated by Taskin and Al, an approach that directly compares negative and positive citations to create a sentiment dictionary may be more fruitful than using a whole corpus of works. So far, we have done some exploratory work focusing on papers that have been retracted (such as paper A, above), from which we have developed two sets of papers: those with negative sentiment when citing the work (perhaps lending weight to evidence towards a retraction) and those citing in a neutral way (including those published before the more critical ones). For example, language classed as negative in papers citing paper A included “appeared inconsistent” whilst neutral language included terms such as “reported” and “compared”.

A sentiment dictionary or corpus for science would allow natural language processing algorithms to distinguish between neutral citations and those with more negative connotations, which may indicate refuting or criticising assertions.
Given we have been able to differentiate between the most frequent words and bi-grams used in the two sets, we think it is possible to build a sentiment analysis corpus for science in a fully automatic way. If developed, such a comprehensive corpus may help the community to highlight areas of scientific debate or concern, and to better understand the impact of a scientific article (such as establishing a standard methodology or a new finding that garners a lot of supporting evidence).
This is what we are now working on. We welcome feedback and contributions to our current work to automatically build a sentiment analysis corpus for science. Please contribute on the GitHub repository, annotate this Labs post or, alternatively, you can email us at innovation [at] elifesciences [dot] org.
Do you have an idea or innovation to share? Send a short outline for a Labs blogpost to innovation@elifesciences.org.
For the latest in innovation, eLife Labs and new open-source tools, sign up for our technology and innovation newsletter. You can also follow @eLifeInnovation on Twitter.
References
Abdulaziz K, Perry JJ, Taljaard M, Émond M, Lee JS, Wilding L, Sirois M-J, Brehaut J. 2018. National Survey of Geriatricians to Define Functional Decline in Elderly People with Minor Trauma. Canadian Geriatrics Journal 19.1: 2-8. doi: https://doi.org/10.5770/cgj.19.192.
Angrosh MA, Cranefield S, Stanger N. 2014. Contextual Information Retrieval in Research Articles: Semantic Publishing Tools for the Research Community. Semantic Web Journal 5(4):261-293. url: https://dl.acm.org/citation.cfm?id=2786113.2786115.
Athar A. 2011. Sentiment Analysis of Citations using Sentence Structure-Based Features. Proceedings of the ACL 2011 Student Session 81-87. url: https://www.aclweb.org/anthology/P11-3015 [PDF].
Athar, A. 2014. Sentiment analysis of scientific citations. University of Cambridge, Computer Laboratory Tech Report. url: http://www.cl.cam.ac.uk/techreports/UCAM-CL-TR-856.pdf [PDF].
Bertin M, Atanassova I, Sugimoto CR, Larivière V. 2016. The linguistic patterns and rhetorical structure of citation context: an approach using n-grams. Scientometrics 109:1417-1434. doi: https://doi.org/10.1007/s11192-016-2134-8.
Di Iorio A, Limpens F, Peroni S, Rotondi A, Tsatsaronis G, Achtsivassilis J. 2018. Coloring citations for scholars: investigating palettes. SAVE-SD 2018 conference submission. url: https://save-sd.github.io/2018/submission/diiorio/index.html.
Fischer N, Schulz C, Stieler K, Hohn O, Lange C, Park SS, Aepfelbacher M. 2010. Xenotropic Murine Leukemia Virus–related Gammaretrovirus in Respiratory Tract. Emerging Infectious Diseases 16(6):1000-1002. doi: https://doi.org/10.3201/eid1606.100066.
Garfield E. 1965. Can citation indexing be automated? National Bureau of Standards Miscellaneous Publication 269:189–192. url: https://citeseerx.ist.psu.edu/viewdoc/summary?doi=10.1.1.78.4421.
Grabitz P, Lazebnik Y, Nicholson J, Rife S. 2017. Science with no fiction: measuring the veracity of scientific reports by citation analysis. bioRxiv 172940. doi: https://doi.org/10.1101/172940.
Greenberg Steven A. 2009. How citation distortions create unfounded authority: analysis of a citation network. BMJ 339:b2680. doi: https://doi.org/10.1136/bmj.b2680.
Lawton MP and Brody EM. 1969. Assessment of Older People: Self-Maintaining and Instrumental Activities of Daily Living. The Gerontologist 9(3.1):179–186. doi: https://doi.org/10.1093/geront/9.3_Part_1.179.
Lombardi VC, Ruscetti FW, Das Gupta J, Pfost MA, Hagen KS, Peterson DL, Ruscetti SK, Bagni RK, Petrow-Sadowski C, Gold B, Dean M, Silverman RH, Mikovits JA. 2009. Detection of an Infectious Retrovirus, XMRV, in Blood Cells of Patients with Chronic Fatigue Syndrome. Science 326(5952):585-589. doi: https://doi.org/10.1126/science.1179052.
Rountree SD, Chan W, Pavlik VN, Darby EJ, Doody RS. 2012. Factors that influence survival in a probable Alzheimer disease cohort. Alzheimer's Research & Therapy 4:16. doi: https://doi.org/10.1186/alzrt119.
Sakuma T, Hué S, Squillace KA, Tonne JM, Blackburn PR, Ohmine S, Thatava T, Towers GJ, Ikeda Y. 2011. No evidence of XMRV in prostate cancer cohorts in the Midwestern United States. Retrovirology 8:23. doi: https://doi.org/10.1186/1742-4690-8-23.
Schäfer I, Hansen H, Schön G, Höfels S, Altiner A, Dahlhaus A, Gensichen J, Riedel-Heller S, Weyerer S, Blank WA, König HH, von dem Knesebeck O, Wegscheider K, Scherer M, van den Bussche H, Wiese B. 2012. The influence of age, gender and socio-economic status on multimorbidity patterns in primary care: first results from the multicare cohort study. BMC Health Services Research 12:89. doi: https://doi.org/10.1186/1472-6963-12-89.
Smith RA. 2010. Contamination of clinical specimens with MLV-encoding nucleic acids: implications for XMRV and other candidate human retroviruses. Retrovirology 7:112. doi: https://doi.org/10.1186/1742-4690-7-112.
Switzer WM, Jia H, Hohn O, Zheng HQ, Tang S, Shankar A, Bannert N, Simmons G, Hendry RM, Falkenberg VR, Reeves WC, Heneine W. 2010. Absence of evidence of Xenotropic Murine Leukemia Virus-related virus infection in persons with Chronic Fatigue Syndrome and healthy controls in the United States. Retrovirology 7:57. doi: https://doi.org/10.1186/1742-4690-7-57.
Taşkin Z and Al U. 2018. A content-based citation analysis study based on text categorization. Scientometrics 114(1):335-337. doi: https://doi.org/10.1007/s11192-017-2560-2.
Teufel S, Siddharthan A, Tidhar D. 2006. Automatic Classification of Citation Function. Proceedings of the 2006 Conference on Empirical Methods in Natural Language Processing 103-110. url: https://dl.acm.org/citation.cfm?id=1610075.1610091.
Tomioka K, Kurumatani N, Hosoi H. 2016. Association Between Social Participation and Instrumental Activities of Daily Living Among Community-Dwelling Older Adults. Journal of Epidemiology 26(10):553-561. doi: https://doi.org/10.2188/jea.JE20150253.
Xu J, Zhang Y, Wu Y, Wang J, Dong X, Xu H. 2015. Citation Sentiment Analysis in Clinical Trial Papers. AMIA Annual Symposium Proceedings 2015:1334–1341. url: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4765697/.
Yu B. 2013. Automated Citation Sentiment Analysis: What Can We Learn from Biomedical Researchers. Proceedings of the 76th ASIS&T Annual Meeting: Beyond the Cloud: Rethinking Information Boundaries 83:1-9. url: http://dl.acm.org/citation.cfm?id=2655780.2655863.




