This post is part of the Innovator Stories Series. At eLife Innovation, we are constantly amazed by open-science ideas and prototypes, but perhaps even more so, we are continuously inspired by the open-source community: its talents, creativity and generosity. In bringing into focus the people behind the projects, we hope that you too will be inspired to make a difference.
By Emmy Tsang
A machine learning engineer by training, Patrice has always been interested in building tools to extract knowledge from the scientific literature, to facilitate the acquisition of new scientific information such that scientists can more easily formulate interesting hypotheses based on existing knowledge.
Most of the scientific literature currently lives in the form of PDFs – a lightweight, universal file format that keeps consistency in text layouts and formatting. The neat and consistent layout of a PDF helps us read and assimilate scientific knowledge. For example, we can easily look at a paper and identify the title and figure captions; the formatting guides us through and helps us navigate the article. However, to a machine, a PDF is a very noisy ASCII file that contains no information on the structure of the text.

Source: Patrice Lopez
Working at the European Patent Office helped Patrice realise that in order to use machines for knowledge extraction, he first needed to pre-process PDFs into machine-friendly formats. Public patent documents are XML files, which are essentially information wrapped in tags, and this allows knowledge extraction algorithms to access and analyse the information stored in these documents. The growing amount and complexity of research, coupled with the increasing sophistication of machines, created the perfect opportunity for us to start using machines to help curate data and extract scientific knowledge, and Patrice quickly realised that to unlock scientific knowledge from PDFs, he needed to create a tool to turn PDFs into XMLs.
This idea gave birth to GROBID, a machine learning library for extracting, parsing and restructuring raw documents such as PDF into structured XML/TEI-encoded documents. GROBID’s parsing and extraction algorithm mainly uses the Wapiti CRF library, which can segment and label sequences of text. In addition to lexical information, GROBID uses the layout information, like the position of the text, or the coordinates, to help characterise text and identify the title, authors, attribution, figure captions, citations and so on. It can also recognise figures and figure references in the rest of the document.
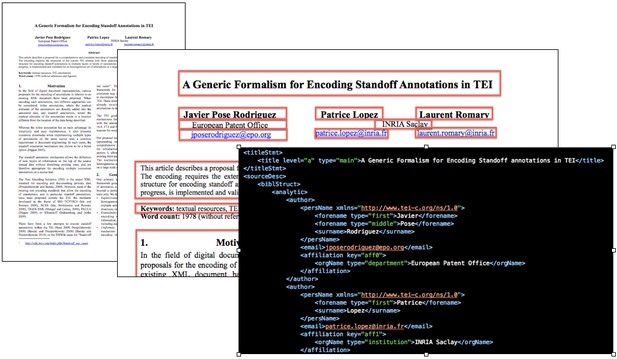
Extracting structured information from PDFs with GROBID. Source: science-miner.com
Patrice’s ambition from the beginning was to create a tool that could process not only a few documents, but the entire corpus of scientific literature. Approximately 2.5m scientific papers are published every year, amounting to terabytes of data, all with different styles and formatting depending on the journal. For GROBID to be scalable, robust and sustainable, Patrice needed to find solutions that will overcome constraints on memory, time and variability in PDF formatting.
Building GROBID in the open
Patrice knew that he needed to develop GROBID openly. First, this means that he can reuse his own work in the future. In previous experiences where he worked on commercial products, he found it frustrating that he could no longer work on a project after he left the company. Developing GROBID open-source has given him the freedom and flexibility to explore and experiment with new technologies and algorithms at his own pace: GROBID can always be more accurate and robust, and implementing different, cutting-edge Natural Language Processing (NLP) and deep learning algorithms and benchmarking them against one another means that the tool is living and constantly improving.
You have to be clear about your motivations, whether it is to learn, or to make an impact.
Second, working on GROBID open-source also allows other developers and technologists to contribute to it. Building a developer’s community around GROBID was not easy at the beginning. As Patrice notes: “It can be frustrating that there’s no feedback, because you’re making the first moves. You have to be clear about your motivations, whether it is to learn, or to make an impact.” While it does mean that time and effort need to be spent on documentation, maintaining issues and pull requests, and generally ensuring the code is accessible, Patrice finds a lot of joy in developing with a community that is dedicated to contributing and sharing, towards driving a positive change.
Patrice also understands that in order to create a tool that is impactful and can serve a wider community, he needs to identify and work with different users. With GROBID being an open tool, technologists developing other platforms can build on top of and contribute back to it, in order to best suit their use cases. Various components of GROBID are now adopted by other research tools such as Mendeley, ResearchGate, the Internet Archive and eLife’s Libero Reviewer. “If you are a researcher, you are very likely to be using GROBID without knowing it,” Patrice said, “and I will not have been able to reach this user base if it were commercial software.”
A recent project built on top of GROBID that Patrice is very excited about involves making books accessible for the visually impaired. It is very difficult for these readers to understand the structure of a page from an eBook, and the necessary tagging of paragraphs, sections and chapters needs to be carried out manually. In this project, led by BrailleNet, GROBID is used to help accelerate and automate this process, ultimately making it easier for visually impaired people to read, work and contribute knowledge.
If you are a researcher, you are very likely to be using GROBID without knowing it.
Knowledge beyond papers
The increasing number and variety of use cases for GROBID drive the software’s continuous evolution. Yet, one interesting dilemma that has arisen is that research papers employing GROBID’s technologies are still citing the single paper that Patrice authored and published about GROBID from 2009. “I think a project is not just code – it’s a lot of documentation, benchmarking and the community,” Patrice commented, “it’s so much more than a paper.” For a machine learning tool development project, to publish is a type of milestone with a date stamp, but a paper captures very little of the richness and dynamics of the project. Along similar lines, citations also reflect only a very small, specific proportion of the tool’s impact. As in Patrice’s case, the current convention to cite a piece of software is to cite the original paper, but often this citation does not refer to the version of the software that was used in the paper, nor does it acknowledge the additional contributors who may have put substantial effort into more recent versions of the software, but who were not authors on the original paper.
To this end, in collaboration with Professor James Howison from The University of Texas at Austin and Impactstory, and supported by a grant from the Sloan Foundation, Patrice is developing an open-source tool that can recognise software mentions in the scientific literature. This tool builds on top of GROBID, and by collecting more accurate open-source software usage data, Patrice and his collaborators hope to better attribute open-source work to reflect the impact of project outputs, so that more researchers will be motivated to develop better and more sustainable open-source tools. “Open-source software is a very valuable research product and important contribution to scientific research, as it contextualises ideas,” Patrice said. “Researchers should receive credit that scales with the usage of the open-source software that they work on.” In the future, he would like to see a tool that can trace both explicit dependencies between software packages and implicit usage mentions in the research literature.
I think a project is not just code – it’s a lot of documentation, benchmarking and the community, it’s so much more than a paper.
Patrice still reads a lot of papers, especially on text mining and NLP, to learn about new models and algorithms that he could use in GROBID and other open-source software. But in building a tool to unlock scientific knowledge from PDFs and papers, he has also come to realise that a lot of knowledge and research activities cannot be captured within a paper. The web has accelerated and added dimensions to research: scientists can now collaborate in real-time on a much larger scale, and data and methods are constantly mutating and improved upon. Software and tool development projects are capable of capturing a much larger proportion of the ongoing research activity and knowledge, and by better acquiring and structuring this information and making it available, we are hopefully moving towards new avenues to assimilate, appreciate and reuse the fruits of scientific research.
#
We welcome comments, questions and feedback. Please annotate publicly on the article or contact us at innovation [at] elifesciences [dot] org.
Do you have an idea or innovation to share? Send a short outline for a Labs blogpost to innovation [at] elifesciences [dot] org.
For the latest in innovation, eLife Labs and new open-source tools, sign up for our technology and innovation newsletter. You can also follow @eLifeInnovation on Twitter.




