Multidimensionality of tree communities structure host-parasitoid networks and their phylogenetic composition
Figures
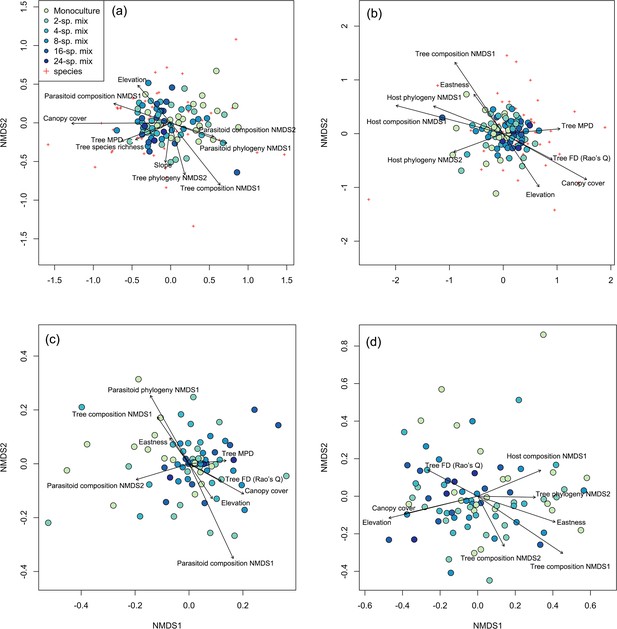
Associations among tree, host, and parasitoid species and phylogenetic composition.
Ordination plot of the nonmetric multidimensional scaling (NMDS) analysis of (a) host species composition, (b) parasitoid species composition, (c) host phylogenetic composition, and (d) parasitoid phylogenetic composition across the study plots (filled circles) in the BEF-China experiment. Stress = 0.23, 0.23, 0.24, and 0.20, respectively. Arrows indicate significant (at p<0.05) correlations of environmental variables with NMDS axis scores. Lengths of arrows are proportional to the strength of the correlations. Red crosses refer to the host or parasitoid species in each community. See Supplementary file 1b–e in the Supplementary Materials for abbreviations and statistical values.
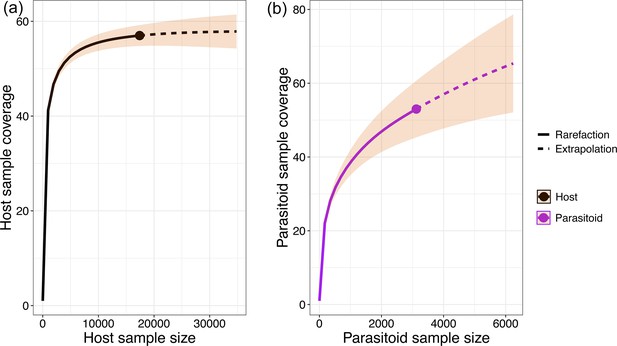
Sampling completeness assessment.
The sample coverage across different sample sizes for (a) hosts and (b) parasitoids.
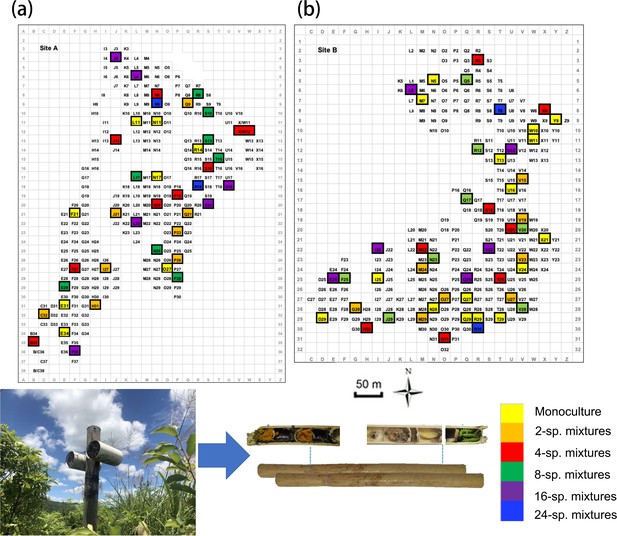
Overview of the study plot distribution along the two experimental tree diversity sites of BEF-China (a: Site A, b: Site B).
Levels of tree species richness indicated by color. Each study plot had a size of 25.8 m × 25.8 m.
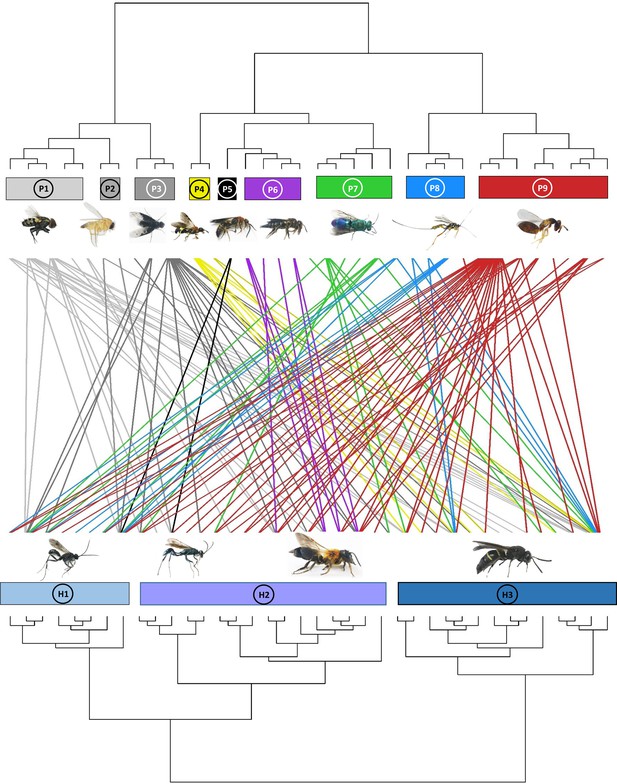
Dendrogram of phylogenetic congruence for the host species (below) and associated parasitoid species (above) recorded in the study.
Each rectangle represents a different superfamily (for host species) or family (for parasitoid species). H1: Pompilidae, H2: Apoidea, H3: Vespidae; P1: Sarcophagidae, P2: Phoridae, P3: Bombyliidae, P4: Trigonalyidae, P5: Mutillidae, P6: Megachilidae, P7: Chrysididae, P8: Ichneumonidae, P9: Chalcidoidea. The trophic network of hosts and parasitoids was nonrandomly structured (parafit test: p=0.032). Host and parasitoid species names are given in Figure 2—figure supplement 1.
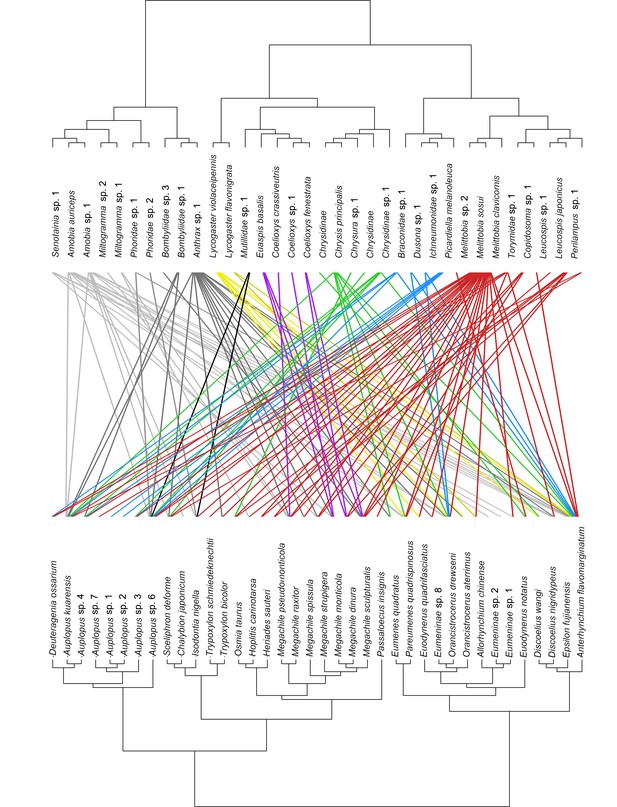
Dendrogram of phylogenetic congruence for the host species (below) and associated parasitoid species (above) recorded in the study, showing the species name for hosts and parasitoids.
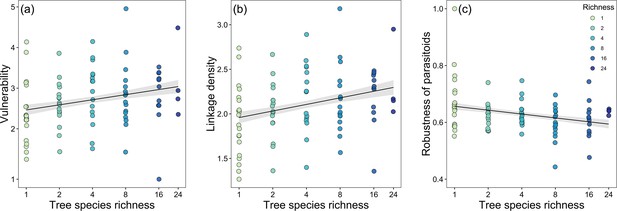
Bivariate relationships between tree species richness and network indices.
Community-level relationships of network between tree species richness and (a) vulnerability, (b) linkage density, and (c) robustness of parasitoids.
Values were adjusted for covariates of the final regression model. Regression lines (with 95% confidence bands) show significant (p<0.05) relationships. Note that axes are on a log scale for tree species richness.
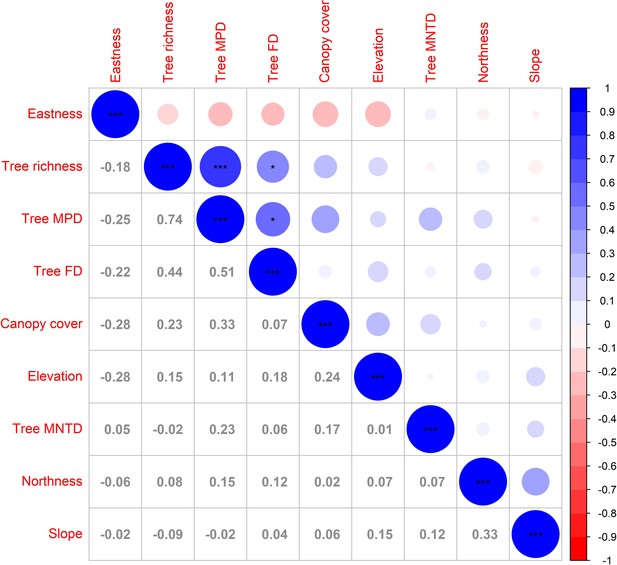
Correlations among the predictors used in the study.
Values and colored circles are Pearson’s correlation coefficients r, significances are indicated by asterisks: ***p<0.001, **p<0.01, *p≤0.05. Tree MPD (tree phylogenetic mean pairwise phylogenetic distance), tree FD (tree functional diversity expressed as Rao’s Q), tree MNTD (tree mean nearest taxon distance).
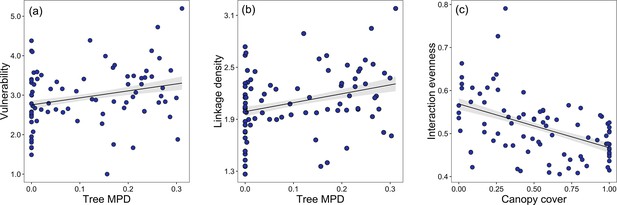
Bivariate relationships between tree MPD, canopy cover and network indices.
Community-level relationships of network between tree phylogenetic mean pairwise distance and (a) vulnerability and (b) linkage density and community-level relationships of network between canopy cover, and (c) interaction evenness.
Values were adjusted for covariates of the final regression model. Regression lines (with 95% confidence bands) show significant (p<0.05) relationships.
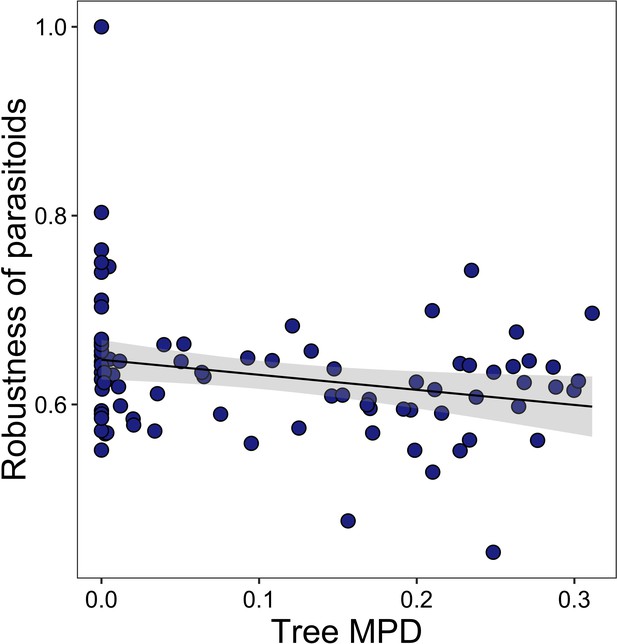
Community-level relationships of networks between tree phylogenetic mean pairwise distance (MPD) and robustness of parasitoids.
Values were adjusted for covariates of the final regression model (i.e., partial residuals shown on y-axis). Regression lines (with 95% confidence bands) show significant (p<0.05) relationships.
Tables
Environmental correlates of dissimilarity matrixes with predictors (nonmetric multidimensional scaling [NMDS] on Morisita-Horn dissimilarity) across the study plots.
Significant p-values are indicated in bold. See Supplementary file 1b–e for the complete information.
| Host species community | Parasitoid species community | Host phylogenetic community | Parasitoid phylogenetic community | |
|---|---|---|---|---|
| Tree phylogeny NMDS1 | 0.225 | 0.422 | 0.386 | 0.274 |
| Tree phylogeny NMDS2 | 0.003 | 0.12 | 0.128 | 0.024 |
| Tree composition NMDS1 | 0.001 | 0.001 | 0.001 | 0.001 |
| Tree composition NMDS2 | 0.604 | 0.418 | 0.433 | 0.031 |
| Canopy cover | 0.001 | 0.001 | 0.001 | 0.004 |
| Tree species richness | 0.035 | 0.122 | 0.100 | 0.094 |
| Elevation | 0.005 | 0.007 | 0.001 | 0.001 |
| Eastness | 0.079 | 0.045 | 0.04 | 0.001 |
| Northness | 0.49 | 0.837 | 0.821 | 0.340 |
| Slope | 0.031 | 0.507 | 0.507 | 0.959 |
| Tree FD (Rao’s Q) | 0.094 | 0.019 | 0.021 | 0.031 |
| Tree MPD | 0.005 | 0.021 | 0.013 | 0.223 |
| Host phylogeny NMDS1 | – | 0.016 | – | 0.584 |
| Host phylogeny NMDS2 | – | 0.027 | – | 0.914 |
| Host composition NMDS1 | – | 0.001 | – | 0.008 |
| Host composition NMDS2 | – | 0.169 | – | 0.138 |
| Parasitoid phylogeny NMDS1 | 0.001 | – | 0.001 | – |
| Parasitoid phylogeny NMDS2 | 0.462 | – | 0.058 | – |
| Parasitoid composition NMDS1 | 0.001 | – | 0.001 | – |
| Parasitoid composition NMDS2 | 0.014 | – | 0.001 | – |
Summary results of linear models for parasitoid generality, host vulnerability, robustness, linkage density, and interaction evenness of host-parasitoid network indices at the community level across the tree species richness gradient.
Standardized parameter estimates (with standard errors, t- and p-values) are shown for the variables retained in the minimal models.
| Est. | SE | t | p | ||
|---|---|---|---|---|---|
| Parasitoid generality | Intercept | 0.176 | 0.016 | 10.96 | <0.001 |
| Canopy cover | 0.033 | 0.016 | 2.03 | 0.046 | |
| Host vulnerability | Intercept | 2.873 | 0.08 | 35.89 | <0.001 |
| Elevation | –0.140 | 0.08 | –1.66 | 0.101 | |
| Tree species richness: Site A | 0.150 | 0.13 | 1.20 | 0.234 | |
| Tree species richness: Site B | 0.230 | 0.11 | 2.12 | 0.037 | |
| Robustness of parasitoids | Intercept | 0.630 | 0.01 | 84.43 | <0.001 |
| Tree species richness: Site A | –0.022 | 0.01 | –1.99 | 0.049 | |
| Tree species richness: Site B | –0.019 | 0.01 | –1.90 | 0.061 | |
| Linkage density | Intercept | 2.038 | 0.04 | 50.99 | <0.001 |
| Elevation | –0.078 | 0.04 | –1.85 | 0.069 | |
| Tree species richness: Site A | 0.106 | 0.06 | 1.70 | 0.094 | |
| Tree species richness: Site B | 0.106 | 0.04 | 2.68 | 0.009 | |
| Intercept | 0.511 | 0.009 | 59.12 | 0.025 | |
| Interaction evenness | Canopy cover | –0.037 | 0.007 | –5.06 | <0.001 |
| Eastness | –0.018 | 0.007 | –2.50 | 0.015 | |
Additional files
-
Supplementary file 1
Supplementary tables were included in this file.
- https://cdn.elifesciences.org/articles/100202/elife-100202-supp1-v2.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/100202/elife-100202-mdarchecklist1-v2.docx





