The lost world of Cuatro Ciénegas Basin, a relictual bacterial niche in a desert oasis
Figures
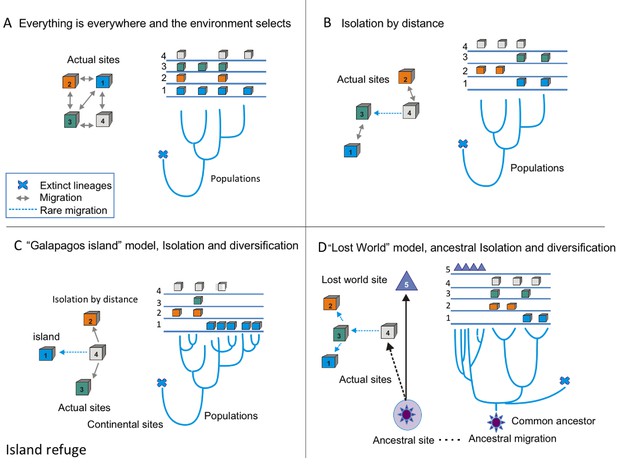
Conceptual frame-work of species diversification.
(A) ‘Everything is everywhere, and the environment selects’ model implies that free migration is only restricted by environmental filtering. Hence, sites 1–4 have the same probability of migration, but some sites, such as one are better than others. In this case, the phylogenetic tree does not reflect the geographic structure; this is common in many cosmopolitan lineages of bacteria and fungi. (B) Model of isolation by distance, this is what occurs in most plants and animal phyla, sites that are closer (1 and 3 or 2 and 4) are more likely to present migration events than sites that far apart, some rare events of migration are allowed (as between 4 and 3). In this case, the branches of the tree reflect the geographic structure. (C) Island model implies that rare events of migration from the source, (4) to an island, such as Galapagos (1). The phylogentic tree reflects adaptive radiation due to isolation. (D) ‘Lost world model’ of ancestral isolation and diversification implies that lineages that were extinct in other places have remained as relictual niches persist in a new site (5). In this case, the ancestral lineages have very long branches that show their ancestral diversification from common ancestors.
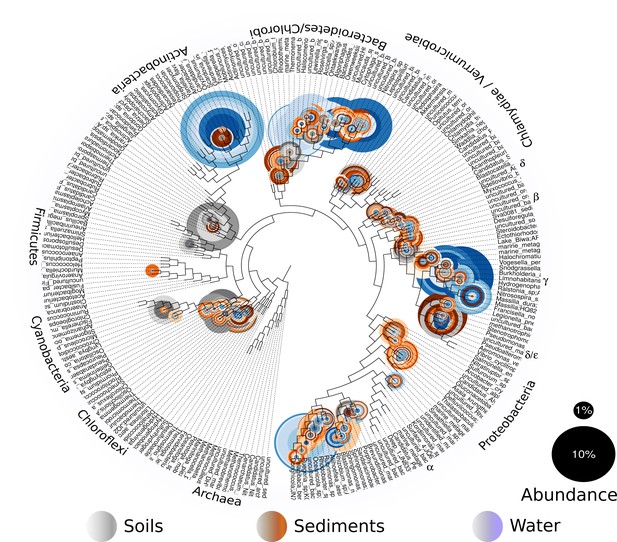
Overall prokaryotic diversity in Churince.
Major phyla abundances in CCB is depicted in bar plots, only the most abundant phyla are shown, but there are 60 phyla present in CCB which are roughly 66.28% of known prokaryotic phyla, in a single location few meters away. Proteobacteria is the most abundant phylum, followed by Bacteroidetes, and Actinobacteria. Some phyla like Planctomycetes, Cyanobacteria, Acidobacteria, Chlorobi, and Firmicutes are more abundant in the sediment and soil-associated samples than in water columns. Each CCB sample is colour-coded according to its origin: blue for water; brown for sediments; and grey for soils.
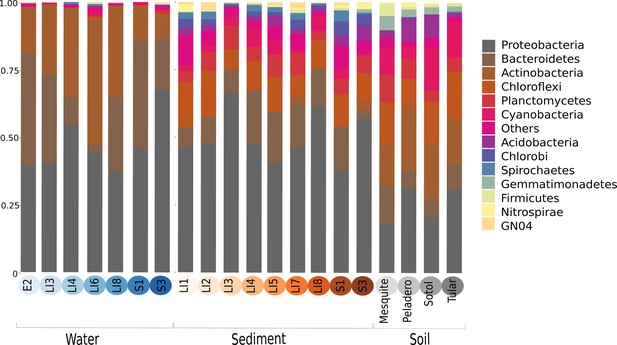
The phylogenetic placement per sampling site shows the names and abundances of the most represented genera within CCB samples.
Even though each site has a particular profile, we can see the aggrupation by type of sample: soil, sediment and water.
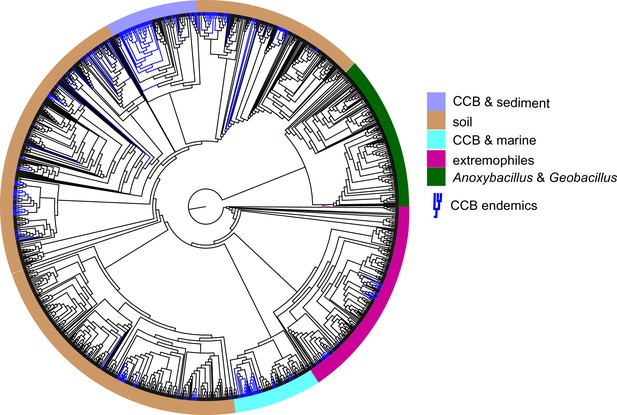
Dendrogram of the 1284 strains of the genus Bacillus reconstructed from 16 s rRNA.
All taxa have a sequence divergence over 97%. Strains found in Cuatro Cienegas Basin (CCB) are denoted in blue. All other strains, including the outgroups, are denoted in light gray. The position of the sediment CCB Bacillus and marine CCB Bacillus lineages within the genus Bacillus are also indicated. The outgroup includes strains of Geobacillus and Anoxybacillus..
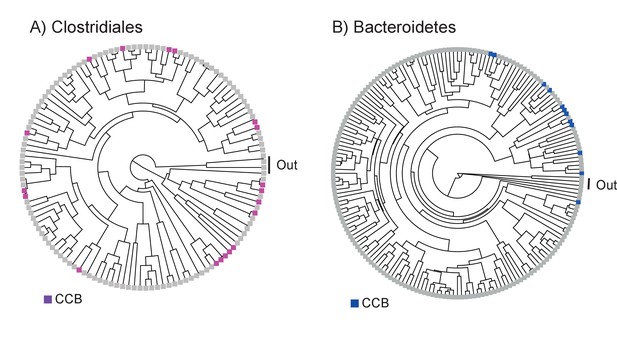
Bayesian phylogenetic trees of the order Clostridiales and the phylum Bacteroidetes reconstructed with 16 s rRNA.
All taxa have a sequence divergence of 97%. Taxa exclusive to CCB are denoted in magenta for Clostridiales and blue for Bacteroidetes. All other taxa, including outgroups, are in light gray. The outgroup in Clostridiales includes Thermoanaerobacter and Symbiobacterium, and for Bacteroidetes the outgroup includes Chlorobium and Prosthecochloris.
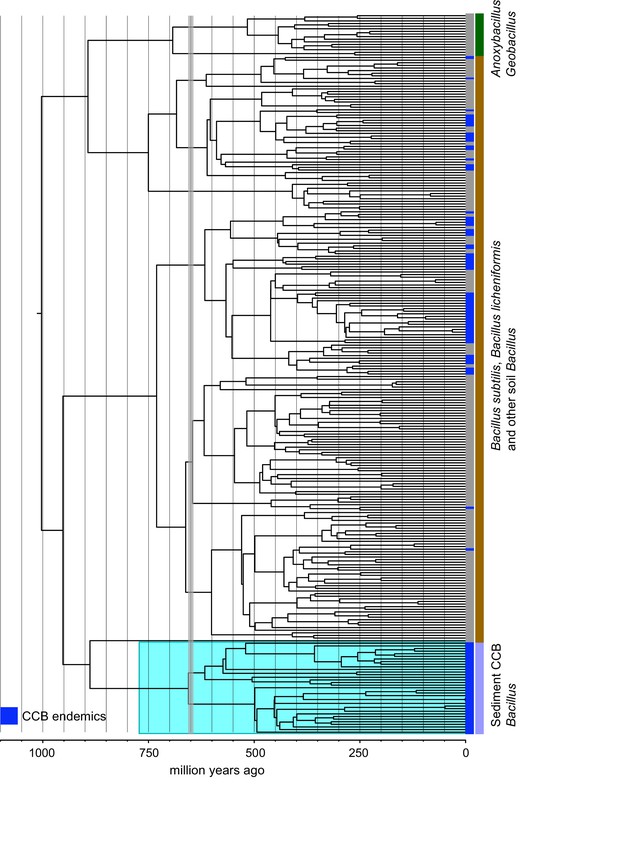
Dated Bayesian phylogeny of soil and sediment Bacillus, including the endemic lineage of sediment CCB Bacillus (highlighted in cyan).
Strains endemic to CCB are denoted in blue. The vertical grey line indicated the date of divergence of sediment CCB Bacillus approximately 655 Ma, in the late Precambrian, during the Cryogenian period.
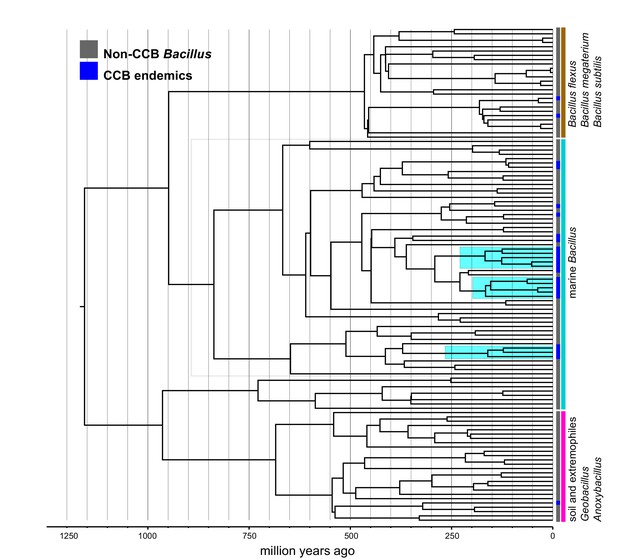
Dated Bayesian phylogeny of marine Bacillus, including endemic lineages from CCB (highlighted in cyan).
The grey line indicates the divergence time of three independent CCB marine strains at around 160 Ma in the Late Jurassic period.
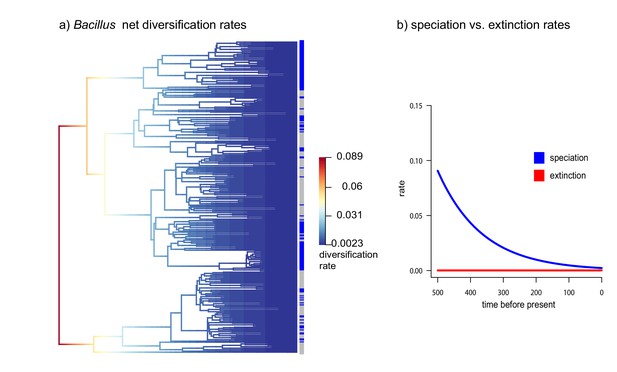
Diversification vs Extinction rates in Bacillus from CCB.
(a) Net diversification rates in soil and sediment Bacillus plotted in a phylogenetic tree. (b) Speciation and extinction rates of soil and sediment Bacillus. Speciation rates lower over time, while extinction rates remain constant.
Additional files
-
Supplementary file 1
Churince’s diversity by site.
- https://doi.org/10.7554/eLife.38278.011
-
Supplementary file 2
Genebank accessions numbers of strains used in this study.
- https://doi.org/10.7554/eLife.38278.012
-
Transparent reporting form
- https://doi.org/10.7554/eLife.38278.013





