Algal-fungal symbiosis leads to photosynthetic mycelium
Figures

Interaction between N.oceanica and M. elongata cells.
(A) Co-cultivation of M. elongata AG77 and N. oceanica (Noc) in flasks for 6 days. Green tissues indicated by the red arrow head are aggregates formed by AG77 mycelium and attached Noc cells. (B) Differential interference contrast micrographs of the green tissues shown in (A). A large number of Noc cells are trapped by AG77 mycelium. (C–F) Scanning electron microscopy images of alga-fungus interaction. (C) Noc cells stick to the fungal mycelium after 6-day co-culture. (D) Noc controls grown in f/2 medium alone have smooth surface. (E) A Noc cell adheres to an AG77 hypha by the outer surface with fibrous extensions, which were exposed after break of the original out layer. Yellow arrows indicate the residues of the out layer. (F) A Noc cell anchored to the AG77 hypha by the fibrous extensions. Red arrows indicate irregular tube-like extensions of the Noc cell wall connected to the surface of fungal cell wall.
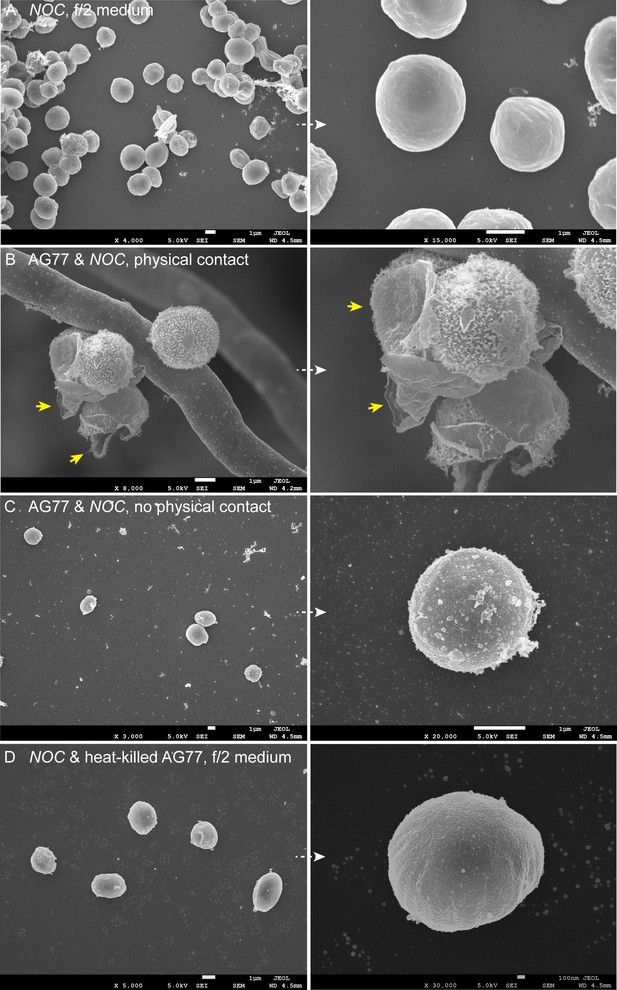
Interaction between N.oceanica cells and M. elongata AG77 hyphae.
Interaction between N. oceanica (Noc) cells and M. elongata AG77 hyphae. (A) Noc controls grown in f/2 medium alone. The cells have smooth surface. (B) Noc cells attached to the fungal hypha after 6-day co-culture. The outer layer is broken, and the fibrous extensions are exposed. The algal outer extensions latch onto the rugged surface of the fungal cell wall. Arrowheads indicate broken pieces from the outer layer after co-culture with the fungi. (C) Free Noc cells co-cultivated with AG77 but not trapped by the mycelium. The Noc cells were filtered with PW200 mesh to remove the mycelium and attached algae. The algal outer layer shows partial damage, but the fibrous extensions are not exposed. (D) Noc cells incubated with heat-killed AG77 medium after 6 days, which have smooth surface similar to the Noc controls in (A).
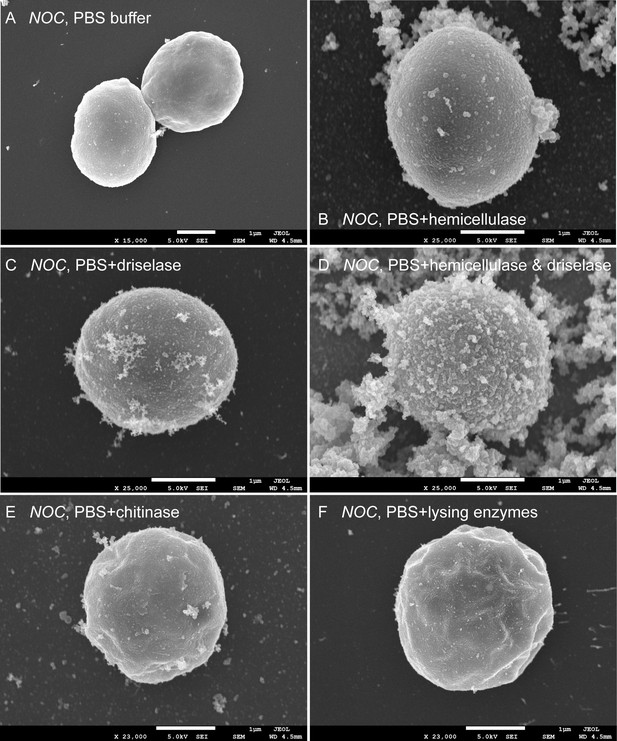
N.oceanica cells were treated with different enzymes to mimic the expose of fibrous extensions after co-culture with fungi.
N. oceanica cells were treated with different enzymes to mimic the exposure of fibrous extensions after co-culture with fungi. (A) Noc cells grown in PBS buffer alone. (B) Noc cells treated with PBS + 4% hemicellulase (mixture of glycolytic enzymes such as xylanase and mananase) for 3 hr at room temperature (RT). (C) Noc cells treated with PBS + 2% driselase (mixture of carbohydrolases including laminarinase, xylanase, and cellulase) for 3 hr at RT. (D) Noc cells treated with PBS + hemicellulase and driselase for 3 hr at RT. The fibrous extensions are visible. (E and F) Noc cells treated with PBS + 1% chitinase or PBS + 1% lysing enzymes (mixture of glucanase, protease, and chitinase) for 6 hr at RT as negative controls.
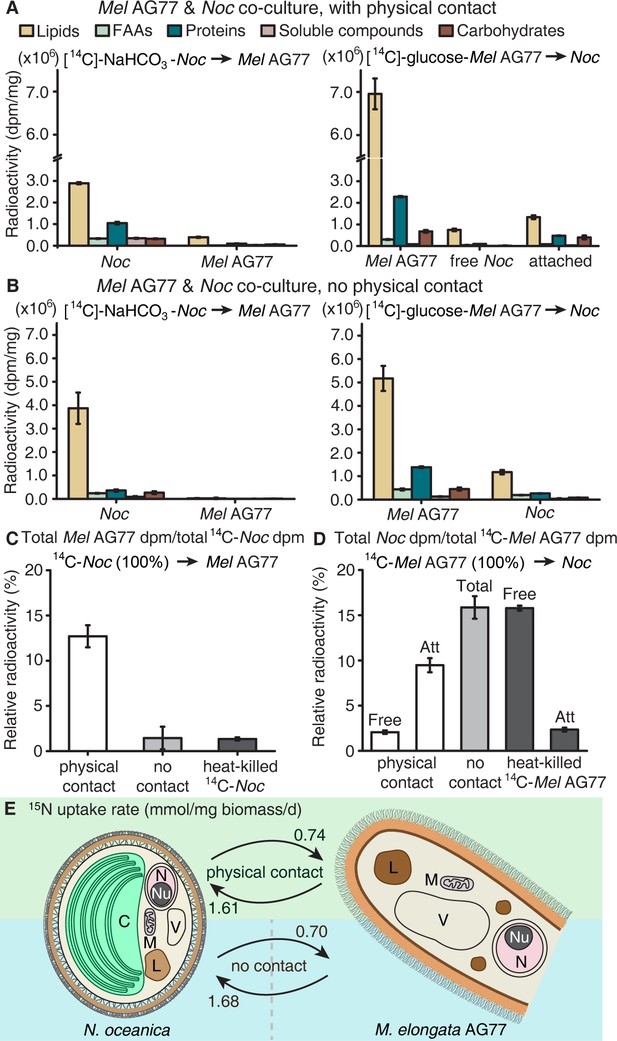
Carbon exchange between N.oceanica and M. elongata AG77.
(A) Carbon (C) transfer from [14C]-sodium bicarbonate (NaHCO3)-labeled N. oceanica (Noc) cells to M. elongata AG77 (Mel AG77, left panel) or from [14C]-glucose-labeled AG77 to Noc cells (right panel) after 7-d co-culture in flasks (with physical contact). Radioactivity of 14C-carbon was determined with a scintillation counter (dpm, radioactive disintegrations per minute) and then normalized to the dry weight of samples (dpm/mg biomass). Free Noc, unbound Noc cells in the supernatant; attached, Noc cells separated from AG77-Noc aggregates by algal cell wall digestion and mesh filtration; FAAs, free amino acids; soluble compounds, supernatant after acetone precipitation of proteins extracted by SDS buffer. Data are presented as the average of three biological replicates with standard deviation (Means ± SD, n = 3). (B) 14C-carbon transfer between Noc and AG77 without physical contact. Algae and fungi were incubated in cell-culture plates with filter-bottom inserts (pore size of 0.4 μm) which separate Noc cells and AG77 mycelium from each other but allow metabolite exchange during co-culture. Error bars indicate SD of three biological replicates (n = 3). (C and D) Relative abundance of 14C-carbon radioactivity in recipient cells compared to 14C-labeled donor cells after 7-d co-culture. (C) AG77 relative to [14C]-NaHCO3-Noc (100%). (D) Noc relative to [14C]-glucose-labeled AG77 (100%). Physical contact, living 14C-labeled cells added to unlabeled cells for co-cultivation in flasks; no contact, samples grown separately in plates with inserts; heat-killed 14C-cells, 14C-labeled Noc or AG77 killed by heat treatment at 65°C for 15 min before the addition to unlabeled cells in flasks. Free, unbound Noc cells in the supernatant; Att, Noc cells attached to AG77 (isolated by algal cell wall digestion and mesh filtration); Total, Noc cells grown separately from AG77 in plates and inserts. Error bars indicate SD of three biological replicates (n = 3). (E) Nitrogen (N) exchange between N. oceanica (Noc) and M. elongata AG77 examined by 15N-labeling experiments. [15N]-potassium nitrate-labeled Noc cells or [15N]-ammonium chloride-labeled AG77 were added to unlabeled AG77 or Noc cells, respectively, for 7-day co-culture in flasks (physical contact) or cell-culture plates with inserts (no physical contact). Algae and fungi were separated and weighed (dry biomass) after the co-culture, and their isotopic composition in Atom% 15N [15N/(15N+14N)100%] and N content (%N) were determined using an elemental analyzer interfaced to an Elementar Isoprime mass spectrometer following standard protocols. The N uptake rate of 15N-Noc-derived N (15N) by AG77 from and that of 15N-AG77-derived N by Noc cells (15N) were calculated based on the Atom% 15N, %N and biomass. C, chloroplast; N, nucleus; Nu, nucleolus; M, mitochondrion; V, vacuole; L, lipid droplet. Values are the average of three biological repeats.
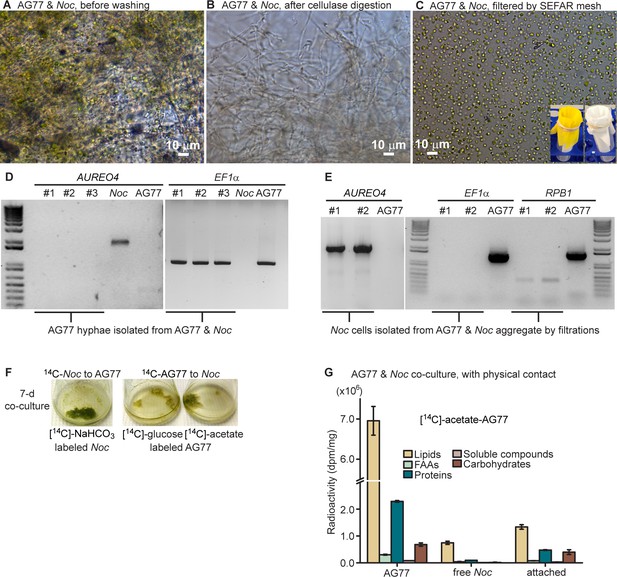
Carbon transfer between N.oceanica and M. elongata AG77 with physical contact.
Carbon transfer between N. oceanica and M. elongata AG77 with physical contact. Separation of N. oceanica and M. elongata AG77 after co-culture and exchange of radioactive carbon ([14C]) between N. oceanica and M. elongata during co-culture. (A) Aggregates of N. oceanica (Noc) cells and AG77 mycelium after 7-day co-culture. (B) Isolated AG77 mycelium from AG77-Noc aggregates after the digestion of hemicellulase and driselase and vortex washes. (C) Isolated Noc cells by mesh filtrations using Accu-Mesh PW200 (yellow mesh, lower right) and then SEFAR NITEX 03-25/14 (white mesh, lower right, mesh opening 25 μm). (D) PCR amplification to examine Noc contamination in isolated AG77 mycelium using primers specific for the gene encoding Aureochrome 4 (AUREO4) in Noc. Primers specific for the fungal gene encoding translation elongation factor EF1α (EF1α) were used as positive controls for fungal tissue. Samples #1 to #3 are biological replicates of isolated fungal mycelium. (E) PCR test for fungal contamination in isolated Noc cells with primers specific for fungal genes encoding EF1α and RNA polymerase RPB1 (RPB1). AUREO4 primers were used as positive controls for the algal cells. Samples #1 and #2 are collected after filtration with PW200 and PW200/NITEX meshes, respectively. (F) Co-culture of N. oceanica (Noc) and M. elongata AG77 in flasks. [14C]-labeled cells were added to unlabeled cells and co-cultured for 7 days; left flask, [14C]-sodium bicarbonate (NaHCO3)-labeled Noc; middle flask, [14C]-glucose-labeled AG77; right flask, [14C]-sodium acetate-labeled AG77. (G) Normalized radioactivity of 14C with respect to the dry weight (dpm/mg). Dpm, radioactive disintegrations per minute; free Noc, unbound cells harvested from the supernatant; attached, Noc cells separated from Noc-AG77 aggregates; FAAs, free amino acids; soluble compounds, supernatant after acetone precipitation of SDS-protein extraction. Values are shown as the average of three biological replicates with standard deviation (Means ± SD, n = 3).
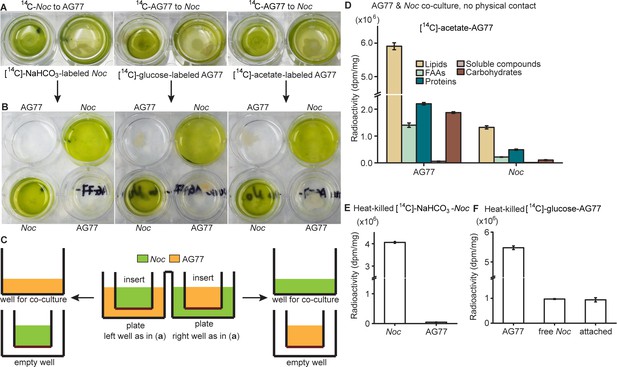
Carbon and nitrogen exchange between N.oceanica and M. elongata AG77 without physical contact.
Carbon and nitrogen exchange between N. oceanica and M. elongata AG77 without physical contact. 14C exchange between N. oceanica and M. elongata AG77 without physical contact using co-culture of N. oceanica (Noc) and M. elongata AG77 in 6-well plates with filter-bottom inserts. (A) Co-cultivation without physical contact. The hydrophilic polytetrafluoroethylene filter (pore size of 0.4 μm) at the bottom of the inserts separates Noc and AG77 during co-culture but allows metabolic exchange between the plate well and insert. [14C]-sodium bicarbonate (NaHCO3)-labeled Noc cells were grown in the plate well or insert while recipient AG77 was grown in the insert or plate well, respectively. Similar incubation conditions were used for [14C]-glucose- or [14C]-sodium acetate-labeled AG77 and recipient Noc. (B) After 7-day co-culture, the inserts were moved to the adjacent empty wells (bottom) for harvesting samples. There was no cross contamination observed between Noc and AG77 samples. (C) Side-view diagram of alga-fungus co-culture (A) and sample harvesting (B) with insert and plate. (D) Detection of 14C transfer from [14C]-sodium acetate-labeled AG77 to recipient Noc. 14C radioactivity (dpm, radioactive disintegrations per minute) was normalized to the dry weight (dpm/mg). FAAs, free amino acids; soluble compounds, supernatant after acetone precipitation of SDS-protein extraction. Values are shown as the average of three biological replicates with standard deviation. (E) 14C transfer between N. oceanica and M. elongata AG77 co-cultured in flasks with physical contact using heat-killed 14C-cells. [14C]-sodium bicarbonate (NaHCO3)-labeled N. oceanica (Noc) cells were killed by incubating them in a water bath at 65°C for 15 min before co-culture with recipient M. elongata AG77 in flasks. 14C radioactivity (dpm, radioactive disintegrations per minute) was measured after separation of Noc and AG77 and then normalized to the dry weight (dpm/mg). Error bars indicate SD, n = 3. (F) Heat-killed [14C]-glucose-labeled AG77 (65°C, 15 min) was added into unlabeled Noc culture and co-cultivated for 7 days in flasks. Normalized radioactivity of 14C is shown as the average of three biological repeats. Free Noc, unbound cells harvested from the supernatant; attached, Noc cells separated from Noc-AG77 aggregates. Nitrogen (N) exchange between N. oceanica (Noc) and M. elongata AG77 examined by 15N-labeling experiments.
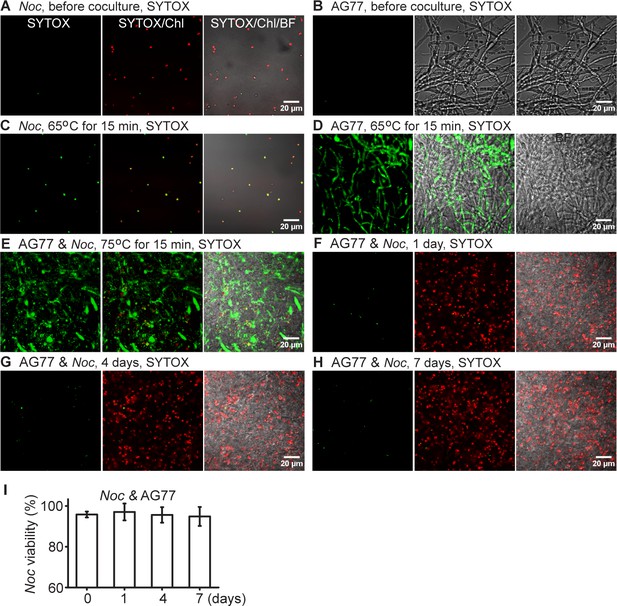
Viability of N.oceanica and M. elongata AG77 during 7-d co-culture.
Viability of N. oceanica and M. elongata AG77 during 7-d co-culture. (A–E) Detection of dead cells in N. oceanica (Noc) mid-log phase culture (A) and M. elongata AG77 grown in PDB medium (B) using SYTOX Green staining. In terms of SYTOX Green-positive controls, Noc (C), AG77 (D) and AG77-Noc aggregates (E) were killed by high temperature in a water bath (65 or 75°C for 15 min) and stained with SYTOX Green. Green fluorescence, SYTOX Green (SYTOX) indicating dead cells; red, Noc chlorophyll fluorescence (Chl); BF, bright field. (F–H) SYTOX Green staining of Noc cells when co-cultured with AG77 for 1 d (F), 4 d (G) and 7 d (H). (I) Viability of Noc cells shown in (F–H) with the 0-d control. Results are calculated from ~2000 cells of six biological repeats with ImageJ. Values are shown as the average of six biological replicates with standard deviation.
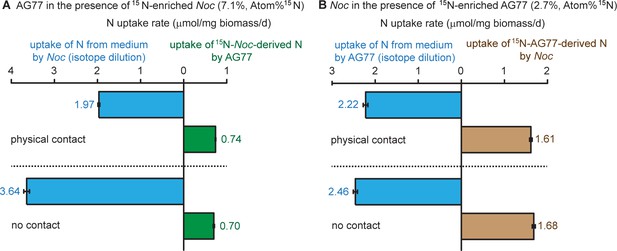
Nitrogen exchange between N.oceanica and M. elongata AG77.
Nitrogen exchange between N. oceanica (Noc) and M. elongata. (A) 15N-potassium nitrate-labeled Noc cells 7.1%, Atom% 15N, 15N/(15N+14N)100% were added to unlabeled AG77 for 7-day co-culture in flasks (physical contact, top) or cell-culture plates with inserts (no physical contact, bottom). Algal and fungal cells were separated and weighed (dry biomass) after the co-culture, and their isotopic composition Atom% 15N [15N/(15N+14N)100%] and N content (%N) were determined using an elemental analyzer interfaced to an Elementar Isoprime mass spectrometer following a standard protocol. The N uptake rates (mmol N/mg biomass/d) of Noc from the medium (medium-N, isotope dilution) and that of AG77 from 15N-Noc-derived N (15N) were calculated based on the Atom% 15N, %N and biomass. Values are shown as the average of three biological replicates with standard deviation, n = 3. (B) Similar analyses were carried out with 15N ammonium chloride-labeled AG77 (2.7%, Atom% 15N) and unlabeled Noc cells to calculate the uptake rate of medium-N by AG77 and that of 15N-AG77-derived N (15N) by Noc cells. Error bars indicate SD, n = 3.
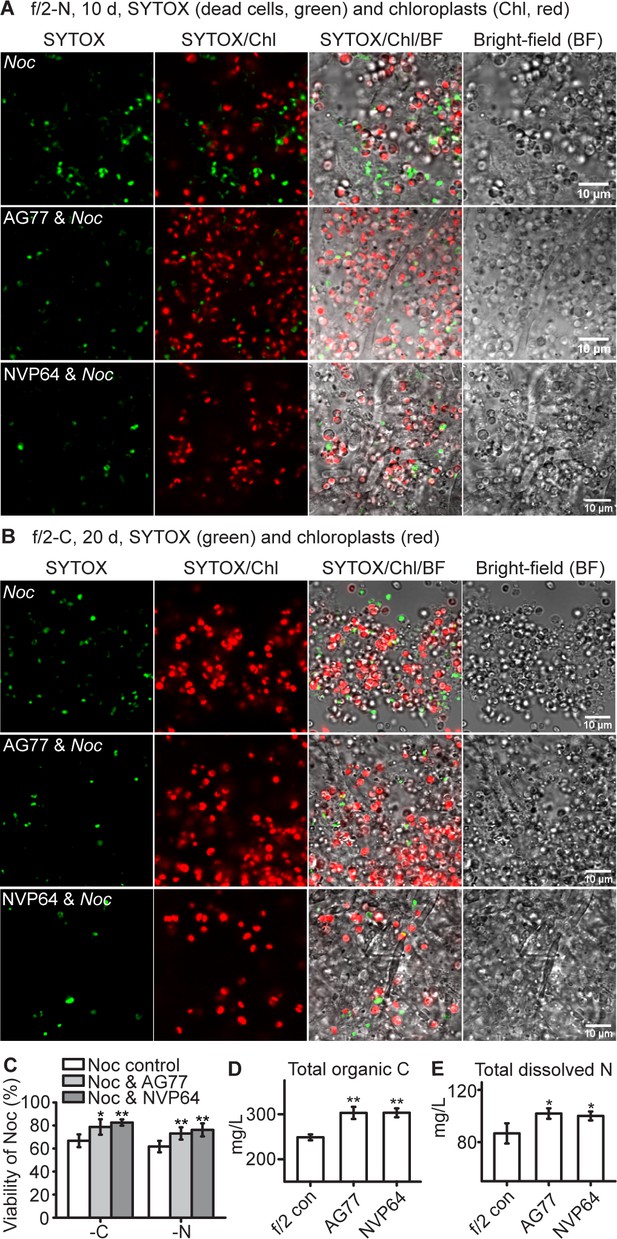
N.oceanica benefits from co-culture with M. elongata.
(A–C) Viability assay of Noc cells and Noc co-cultured with AG77 under nitrogen (-N, (A) and carbon (-C, (B) deprivation. Dead Noc cells were indicated by SYTOX Green staining (green fluorescence). Red, Noc chlorophyll fluorescence. (C) Viability of nutrient-deprived Noc cells increased when co-cultured with two different M. elongata strains, AG77 and NVP64. Results are calculated from 1000 to 5000 cells of five biological repeats with ImageJ. Asterisks indicate significant differences compared to the Noc control as determined by Student’s t test (*p≤0.05, **p≤0.01; Means ± SD, n = 5). (D and E) Total organic C and dissolved N measurements in the buffer of 18-day fungal cultures of M. elongata strains AG77 and NVP64 compared to the f/2 medium control (f/2 con). Fungal cells were removed by 0.22-μm filters. Data are presented as the average of four biological replicates and asterisks indicate significant differences compared to the f/2 medium control as determined by Student’s t test. Means ± SD, n = 4. *p≤0.05, **p≤0.01.
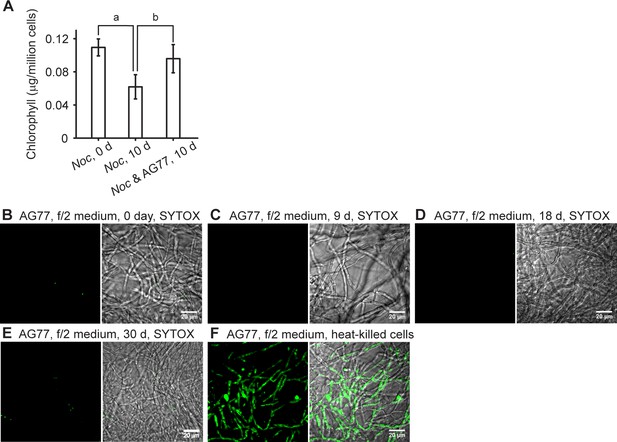
N. oceanica and M. elongata under stresses.
N. oceanica and M. elongata under stresses. (A) N. oceanica (Noc) benefits from co-culture with M. elongata following prolonged cultivation. Stationary-phase Noc cells (0 day control) were kept growing alone or f/2-washed and blot-dried AG77 mycelium were added for 10-day-prolonged incubation. The algae control and unbound algae of the co-culture were used for chlorophyll measurement. Significant chlorophyll degradation was observed in the Noc-alone culture but not in the co-cultured cells. Letter a indicates significant difference compared to the 0 day Noc control; letter b indicates significant difference compared to the 10 days Noc-alone cells (p≤0.05). Values are shown as the average of five biological replicates with standard deviation. (B–F) Viability assay of M. elongata AG77 grown in f/2 medium. Representative confocal micrographs for the detection of dead cells in M. elongata AG77 incubated in f/2 medium for 0 (B), 9 (C), 18 (D) and 30 days (E) using SYTOX Green staining (green fluorescence). Very few dead cells were observed at or before 18-day incubation, which is the same period as for the samples used for organic carbon and dissolved nitrogen measurements in Figure 3D and E. More dead cells were found at 30 days (E), but this number is still very low compared to the heat-killed control (F). Left panel, SYTOX Green fluorescence; right panel, SYTOX/bright field.
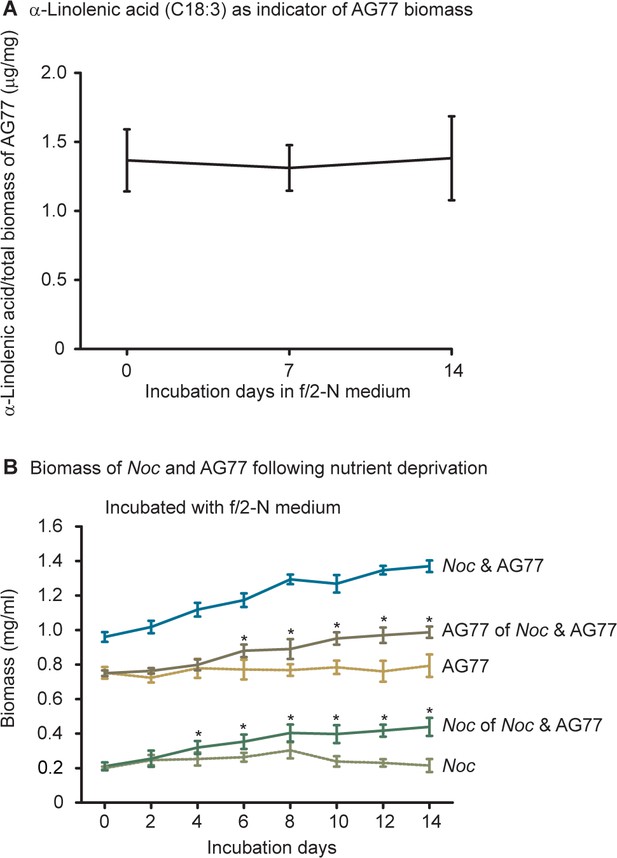
N.oceanica and M. elongata AG77 benefit from each other under nutrient starvation.
N. oceanica and M. elongata AG77 benefit from each other under nutrient starvation. (A) Biomass calculation of M. elongata using linolenic acid (C18:3) as a biomarker, a fatty acid present in AG77 but not in N. oceanica (Noc). The C18:3 composition in total biomass was consistent following the incubation in N-deprived f/2 medium. Values are shown as the average of four biological replicates with standard deviation, n = 4. (B) Dry weight measurement of Noc and AG77 following nutrient starvation. Three groups of samples (Noc, AG77, and Noc and AG77) were washed and incubated in N-deprived f/2 medium (f/2 N) for up to 14 days and were harvested by centrifugation for further fatty acid and biomass measurements at indicated days. Total lipid of Noc-AG77 aggregates was extracted and the cell lysate was dried for the total biomass of Noc and AG77. C18:3 fatty acid methyl esters were determined by gas chromatography and the biomass of AG77 was calculated with C18:3 as a proxy to quantify the fungal biomass taking into account that the C18:3 composition in the fungal biomass was consistent following the incubation in N-deprived f/2 medium. The biomass of Noc in co-culture with AG77 was obtained by subtraction of the AG77 biomass. Noc and AG77 grown by themselves were used as negative controls that did not have obvious increase in biomass following nutrient starvation. Asterisks indicate significant differences (*p≤0.05) between AG77 and AG77 of Noc and AG77 or between Noc and Noc of Noc and AG77 as determined by Student’s t test. Data are presented as the average of four biological replicates with error bars indicating SD (n = 4).
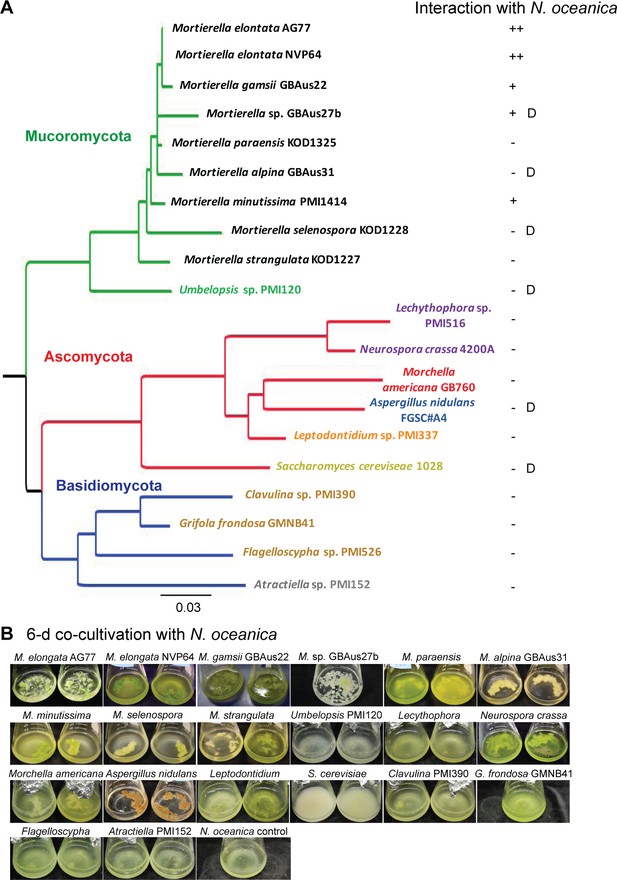
Screening of fungal isolates from diverse clades for intensive interaction with N.oceanica.
Screening of fungal isolates from diverse clades for intensive interaction with N. oceanica. (A) Phylogram based on partial 28S ribosomal DNA sequence data shows the phylogenetic diversity of fungi tested through interaction studies with N. oceanica. Phylogenetic analysis was carried out using the neighbor joining optimization criterion and major groupings are congruent with current understanding of the fungal phylogeny. Phyla are distinguished by branch colors: Mucoromycota (green), Ascomycota (red), Basidiomycota (blue). Taxa are colored in respect to the Order that they belong to: Mortierellales (black), Umbelopsidales (green), Helotiales (orange), Pezizales (red), Eurotiales (blue), Sordariales (purple), Saccharomycetales (yellow), Atractiellales (gray) and Agaricales (brown). Interactions between N. oceanica and the fungi were evaluated after 6-day co-cultivation briefly based on the amount of cells attached to the fungal mycelia (green dots and pieces): ++, obvious green pieces such as AG77 and NVP64; +, relatively fewer green dots such as GBAus27b; -, no green cells observed while the culture is green such as M. paraensis; D, algal cells are dead after the co-culture, such as for GBAus31. (B) Images of N. oceanica cells co-cultured with different fungal species in flasks containing f/2 medium for 6 days.

Scanning electron microscopy of co-cultures of N.oceanica with two fungal strains that did not trap N. oceanica cells.
Scanning electron microscopy of co-cultures of N. oceanica (Noc) with two fungal strains that did not trap Noc cells. (A) Noc and Clavulina PMI390 in f/2 medium for 6 days. A few Noc cells were found attached to the hyphae, which has a smooth surface (right panel). (B) Noc and Morchella Americana GB760 in f/2 medium for 6 days. Noc cells were hardly found in the mycelium. The right panel shows a free Noc cell in the co-culture.
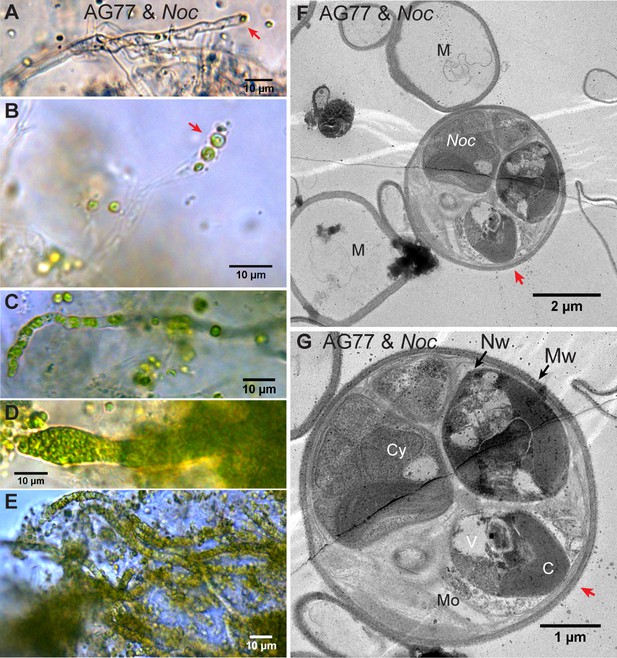
Intracellular localization of long-term co-cultured N.oceanica within M. elongata AG77 hyphae.
(A–E) DIC images of AG77 ‘green hyphae’ with N. oceanica (Noc) cells inside. The red arrow heads indicate Noc cells at the tip region of the hypha. (B and C) AG77 and Noc co-cultured for ~1 month. (C–E) AG77 and Noc co-cultured over 2 months. (F and G) Transmission electron microscope (TEM) images of increasing magnification showing a cross-section of AG77 mycelium containing a cluster of Noc cells. AG77 and Noc were co-cultured for ~1 month. Red arrowheads indicate the same position. M, mycelium; Nw, Noc cell wall; Mw, Mortierella cell wall; Cy, cytoplasm; V, vacuole C, chloroplast; Mo, Mortierella organelles.
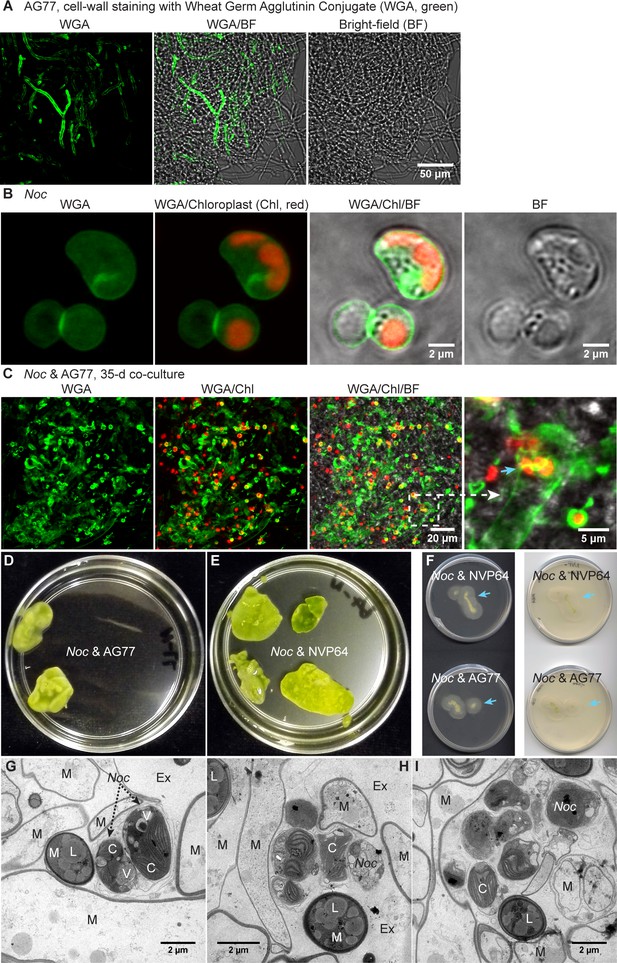
Interaction between N. oceanica and M. elongata AG77 during long-term co-culture by confocal and transmission electron microscopy.
Interaction between N. oceanica and M. elongata AG77 during long-term co-culture by confocal and transmission electron microscopy. (A–C) Confocal microscopy suggesting localization of N. oceanica cells inside M. elongata AG77 hyphae after long-term co-culture. Cell-wall staining of N. oceanica (Noc) and M. elongata AG77 using Wheat Germ Agglutinin Conjugate (WGA, green fluorescence). Confocal laser scanning microscopy with WGA staining allowed us to observe cell structure and symbiosis of living algal and fungal cells inside the AG77-Noc aggregates. (A) AG77 control. (B) WGA stained Noc cells. Red, Noc chlorophyll fluorescence. (C) After co-culture, a cluster of Noc cells is likely growing inside AG77 hyphae as indicated by the blue arrowhead. (D and E) Algae-fungi aggregates after 1 month co-culture. The samples were rinsed in PBS buffer and then used for transmission electron microscopy (TEM). (F) One-month-old algae-fungi aggregates were transferred to fresh PDB/2 plates for overnight incubation. Arrow heads indicate new growth of the fungal mycelium. (G–I) TEM images showing interaction between N. oceanica and M. elongata AG77 during long-term co-culture. Noc cells trapped and surrounded by fungal mycelium. M, mycelium; L, lipid droplet; Ex, extracellular space. C, chloroplast; V, vacuole; Cy, cytoplasm.
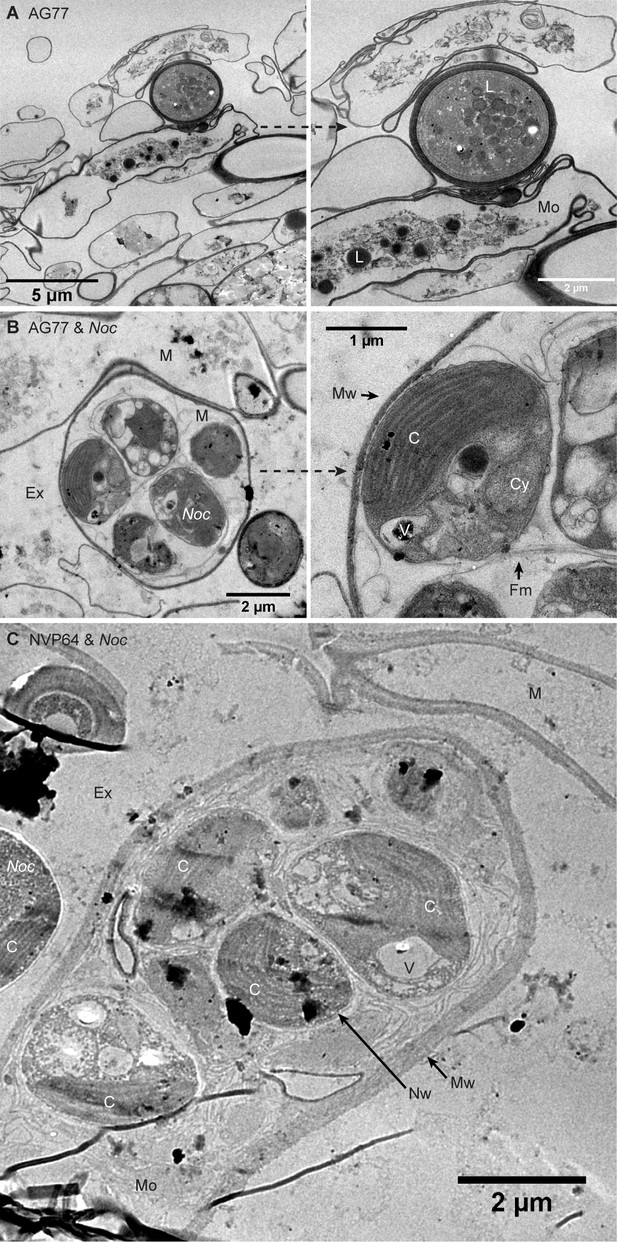
N. oceanica cells inside M. elongata mycelium after long-term co-culture.
N. oceanica cells inside M. elongata mycelium after long-term co-culture. (A and B) TEM images showing N. oceanica (Noc) cells inside the mycelium of M. elongata AG77. M, mycelium; Ex, extracellular space; Mw, Mortierella cell wall; Nw, Noc cell wall; C, chloroplast; Cy, cytoplasm; V, vacuole; L, lipid droplet. (C) Noc cells within a M. elongata NVP64 mycelium. Mo, Mortierella organelles.
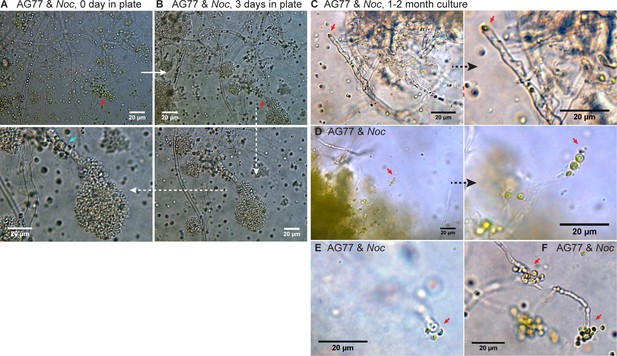
Origin of N.oceanica within M. elongata AG77.
Origin of N. oceanica within M. elongata AG77. (A) A differential interference contrast (DIC) micrograph of co-cultured N. oceanica (Noc) and M. elongata AG77 using a DIC microscope (Leica DMi8). After 35-day co-culture in flasks, AG77-Noc aggregates were transferred to 35-mm-microwell dish (glass top and bottom, MatTek) containing semisolid medium (f/2 medium supplemented with 0.25% low-gelling-temperature agarose and 10% PDB) to investigate the establishment of the Noc endosymbiosis in AG77. The red arrowhead indicates a hypha coated by Noc cells around the hyphal tip. (B) After 3-day incubation in semisolid medium, the same group of Noc and AG77 cells formed a ‘green hypha’ (with Noc cells inside) as indicated by the red arrowhead. Noc cells surrounding the hypha kept growing and dividing into a clavate formation because of the solid medium, which was not observed in the liquid co-culture. In the enlarged clavate region, the cyan arrowhead points to Noc cells inside the fungal hypha. (C–F) Interaction between N. oceanica and M. elongata AG77 at the hyphal tip. DIC micrographs of N. oceanica (Noc) and M. elongata AG77 aggregates co-cultured in flasks containing f/2 medium for 1 to 2 months. Samples were observed in glass top and bottom Petri dishes using a Leica DMi8 DIC microscope. (C) A Noc cell attached to the tip of a hypha as indicated by red arrowheads. (D) Several Noc cells inside an AG77 hypha at the tip area as indicated by red arrowheads at two magnifications. (E and F) Interaction between Noc cells and AG77 at the hyphal tip.

Presence of N.oceanica in M. elongata AG77.
Presence of N. oceanica in M. elongata AG77. DIC micrographs of N. oceanica (Noc) and M. elongata AG77 aggregates co-cultivated in Petri dishes containing semisolid medium (f/2 medium supplemented with 0.25% low-gelling-temperature agarose and 10% PDB). AG77-Noc aggregates from 1 to 2 months co-culture in flasks were transferred to the Petri dish and incubated for 1 to 2 weeks. A large number of green hyphae were observed during the incubation, which had clavate enlargements and aggregations of Noc cells at the hyphal tips. Red arrowheads indicate tip area of the green hyphae.
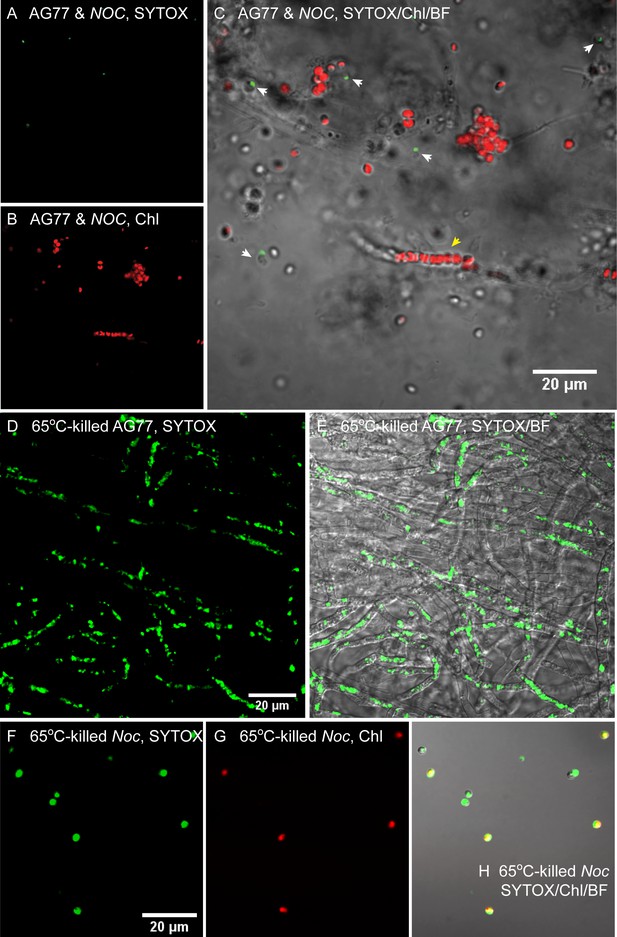
Viability assay of green hyphae.
Viability assay of green hyphae. (A–C) A representative piece of green hyphae stained by SYTOX green to detect dead cells. N. oceanica (Noc) and M. elongata AG77 were co-cultured for 1–2 months before the staining and confocal microscopy. Green, SYTOX in dead cells (A); red, chlorophyll autofluorescence (Chl, (B); overlay of SYTOX, Chl, and bright field (BF, (C). The yellow arrowhead indicates a green hypha containing a group of Noc cells. White arrowheads indicate dead Noc cells around fungal hyphae. (D and E) SYTOX green staining of heat-killed AG77. (F–H) SYTOX green staining of heat-killed Noc. The same microscope settings were used for the heat-killed controls, which show much higher signal than the cells in (A–C).
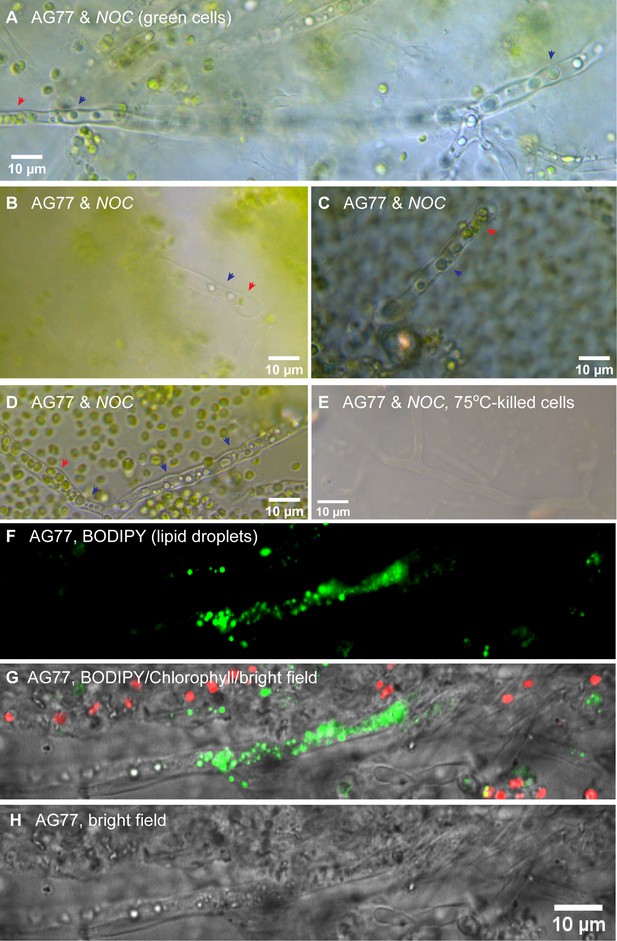
Light microscopy of green hyphae showing the coexistence of N.oceanica and fungal organelles inside hyphae.
Light microscopy of green hyphae showing the coexistence of N. oceanica and fungal organelles inside hyphae. (A–D) Green hyphae after 1–2 month co-culture of N. oceanica (Noc) and M. elongata AG77. Red arrow heads indicate Noc cells (green) inside hyphae. Black arrow heads indicate lipid droplets (homogeneous blue/green) in fungal hyphae. (E) Heat-killed controls. (F–H) BODIPY staining (green) of AG77 and Noc cells showing lipid droplets in fungal hyphae. Red, chlorophyll autofluorescence. Noc cells start to accumulate lipid droplets when they are under stress such as nitrogen deprivation.
Videos
M. elongata AG77 mycelia (~2 days in PDB), N. oceanica (Noc, 5 days in f/2) or AG77-Noc aggregates (7-d co-culture) were washed three times with phosphate-buffered saline (PBS, pH7.0–7.2, Life Technologies) and incubated in flasks containing PBS for ~2 days.
Samples were then transferred to 35-mm-microwell dishes (glass top and bottom, MatTek) containing semisolid medium (PBS supplemented with 0.25% low-gelling-temperature agarose). The growth of samples was recorded by time-lapse photography (every 20 min for 4 days) using a Leica DMi8 inverted microscope with DIC and time-lapse function. Resultant images were used to create the movie with video-editing software (VideoStudio X9, Corel) to compare the growth of AG77 with or without symbiotic algal cells in nutrient-limited PBS buffer. Only hyphae of AG77-Noc aggregates kept growing, indicating that AG77 benefits from the co-culture with Noc cells.
Animation of 3D z-stacks of N. oceanica (Noc) or M. elongata AG77-Noc aggregates (35-day co-culture) stained by Wheat Germ Agglutinin Conjugate (WGA) and observed with a confocal laser scanning microscope (FluoView 1000, Olympus).
Green, WGA fluorescence indicates cell wall of Noc and AG77; red, Noc chlorophyll fluorescence.
Videos recorded with a Leica DMi8 inverted microscope to show the morphology of green hyphae in AG77-Noc aggregates (co-cultured over 2 months).
https://doi.org/10.7554/eLife.47815.025N. oceanica (Noc) cells internalized within a M. elongata AG77 hypha recorded by time-lapse photography (every 10 min for 6 d), showing several Noc cells growing and dividing within the hypha.
https://doi.org/10.7554/eLife.47815.026Light microscope video to show the N. oceanica (Noc) cells inside a M. elongata AG77 hypha.
The green Noc cells are surrounded by fungal organelles such as lipid droplets (blue green/gray) that are presented in living hypha.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain (Nannochloropsis oceanica CCMP1779) | Noc | Provasoli-Guillard National Center for Culture of Marine Phytoplankton | CCAP211/46 | Kuwait Institute for Scientific Research |
| Strain (Mortierella elongata AG77) | AG77/Mel AG77 | Uehling et al., 2017 | North Carolina | |
| Strain (Mortierella elongata NVP64) | NVP64 | Uehling et al., 2017 | Michigan | |
| Strain (Mycoavidus cysteinexigens) | M. cysteinexigens | Uehling et al., 2017 | ||
| Commercial assay or kit | SYTOX Green | Thermo Fisher Scientific | R37168 | |
| Commercial assay or kit | BODIPY 493/503 | Thermo Fisher Scientific | D3922 | |
| Commercial assay or kit | Wheat Germ Agglutinin Conjugate Alexa Fluor 488 | Thermo Fisher Scientific | W11261 | |
| Commercial assay or kit | resin Epon/Araldite mixture | Electron Microscopy Sciences | 13940 | |
| Chemical compound, drug | [14C]-sodium bicarbonate | American Radiolabeled Chemicals | ARC 0138 C-1 mCi | |
| Chemical compound, drug | [14C]-D-glucose | Moravek Biochemicals | MC144W | |
| Chemical compound, drug | [14C]-sodium acetate | American Radiolabeled Chemicals | ARC 0101A | |
| Chemical compound, drug | [15N]-ammonium chloride | Sigma-Aldrich | 299251 | |
| Software, algorithm | VideoStudio X9 | VideoStudio | X9 |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.47815.028





