Science Forum: Wikidata as a knowledge graph for the life sciences
Figures
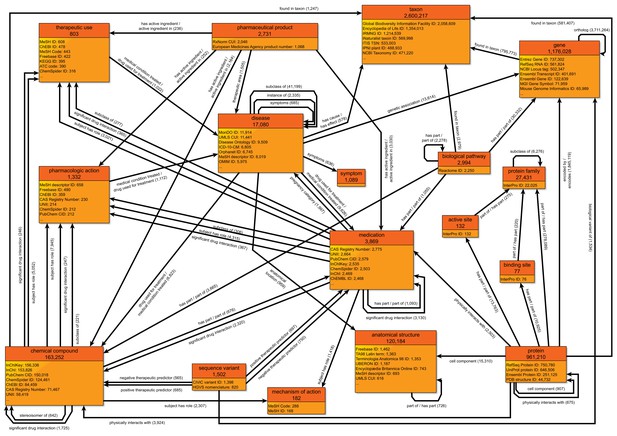
A simplified class-level diagram of the Wikidata knowledge graph for biomedical entities.
Each box represents one type of biomedical entity. The header displays the name of that entity type (e.g., pharmaceutical product) and the number of Wikidata items for that entity type. The lower portion of each box displays a partial listing of attributes about each entity type and the number of Wikidata items for each attribute. Edges between boxes represent the number of Wikidata statements corresponding to each combination of subject type, predicate, and object type. For example, there are 1505 statements with 'pharmaceutical product' as the subject type, 'therapeutic area' as the predicate, and 'disease' as the object type. For clarity, edges for reciprocal relationships (e.g., 'has part' and 'part of') are combined into a single edge, and scientific articles (which are widely cited in statement references) have been omitted. All counts of Wikidata items are current as of September 2019. The most common data sources cited as references are available in Figure 1—source data 1. Data are generated using the code in https://github.com/SuLab/genewikiworld (archived at Mayers et al., 2020). A more complete version of this graph diagram can be found at https://commons.wikimedia.org/wiki/File:Biomedical_Knowledge_Graph_in_Wikidata.svg.
-
Figure 1—source data 1
Most frequent data sources cited as references for the biomedical subset of the Wikidata knowledge graph shown in Figure 1.
- https://cdn.elifesciences.org/articles/52614/elife-52614-fig1-data1-v1.csv
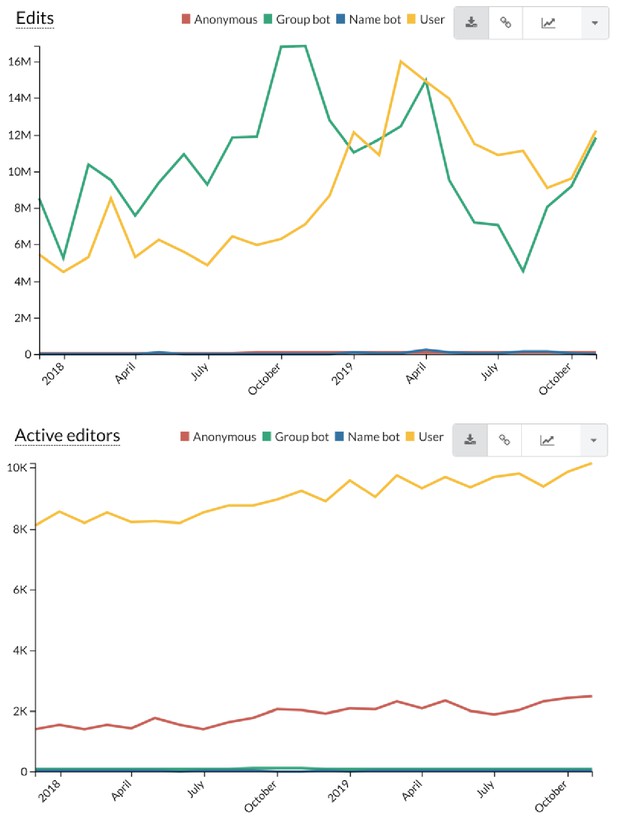
Trends in Wikidata edits.
Wikidata edits are categorized into four categories: anonymous edits with no user account ('anonymous'), edits from formally registered bots ('group bot'), edits from user accounts that are presumed to be bots based on the user account name ('name bot'), and all other edits from registered, logged-in users. The top graph shows that Wikidata receives substantial contributions from both automated bots and individual users. While the overall number of edits is relatively balanced between these two groups, the lower graph shows that the number of user accounts is much higher than the number of automated bot accounts. Statistics are shown for the periods between December 2017 through December 2019. More statistics are available at https://stats.wikimedia.org/v2/#/wikidata.org.
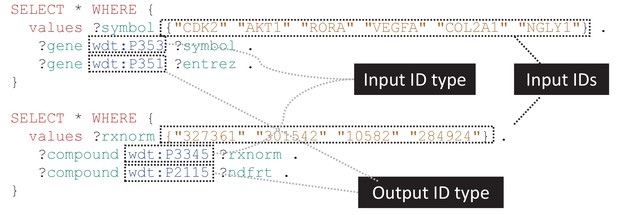
Generalizable SPARQL template for identifier translation.
SPARQL is the primary query language for accessing Wikidata content. These simple SPARQL examples show how identifiers of any biological type can easily be translated using SPARQL queries. The top query demonstrates the translation of a small list of gene symbols (wdt:P353) to Entrez Gene IDs (wdt:P351), while the bottom example shows conversion of RxNorm concept IDs (wdt:P3345) to NDF-RT IDs (wdt:P2115). These queries can be submitted to the Wikidata Query Service (WDQS; https://query.wikidata.org/) to get real-time results. Translation to and from a wide variety of identifier types can be performed using slight modifications on these templates, and relatively simple extensions of these queries can filter mappings based on the statement references and/or qualifiers. A full list of Wikidata properties can be found at https://www.wikidata.org/wiki/Special:ListProperties. Note that for translating a large number of identifiers, it is often more efficient to perform a SPARQL query to retrieve all mappings and then perform additional filtering locally.
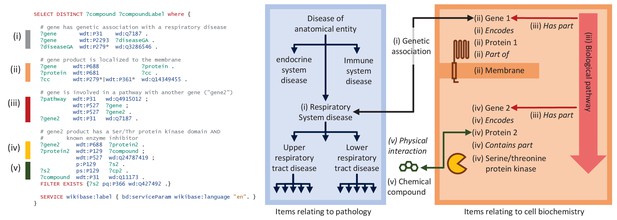
A representative SPARQL query that integrates data from multiple data resources and annotation types.
This example integrative query incorporates data on genetic associations to disease, Gene Ontology annotations for cellular compartment, protein target information for compounds, pathway data, and protein domain information. Specifically, this query (depicted schematically at right) retrieves genes that are (i) associated with a respiratory system disease, (ii) that encode a membrane-bound protein, and (iii) that sit within the same biochemical pathway as (iv) a second gene encoding a protein with a serine-threonine kinase domain and (v) a known inhibitor, and reports a list of those inhibitors. Aspects related to Disease Ontology in blue; aspects related to biochemistry in red/orange; aspects related to chemistry in green. Properties are shown in italics. Real-time query results can be viewed at https://w.wiki/6pZ.
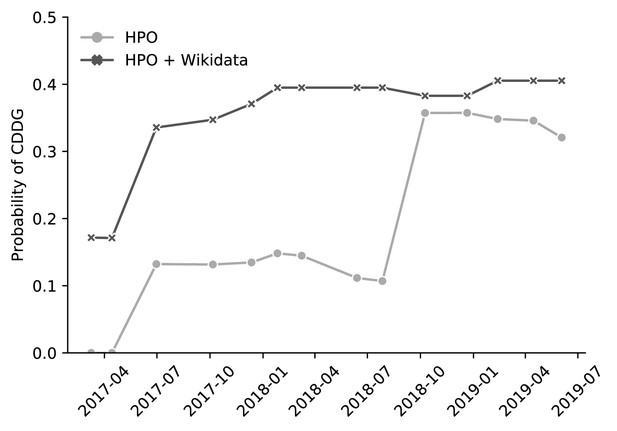
BOQA analysis of suspected cases of the disease Congenital Disorder of Deglycosylation (CDDG).
We used an algorithm called BOQA to rank potential diagnoses based on clinical phenotypes. Here, clinical phenotypes from two cases of suspected CDDG patients were extracted from a published case report (Caglayan et al., 2015). These phenotypes were run through BOQA using phenotype-disease annotations from the Human Phenotype Ontology (HPO) alone, or from a combination of HPO and Wikidata. This analysis was tested using several versions of disease-phenotype annotations (shown along the x-axis). The probability score for CDDG is reported on the y-axis. These results demonstrate that the inclusion of Wikidata-based disease-phenotype annotations would have significantly improved the diagnosis predictions from BOQA at earlier time points prior to their official inclusion in the HPO annotation file. Details of this analysis can be found at https://github.com/SuLab/Wikidata-phenomizer (archived at Tu et al., 2020).
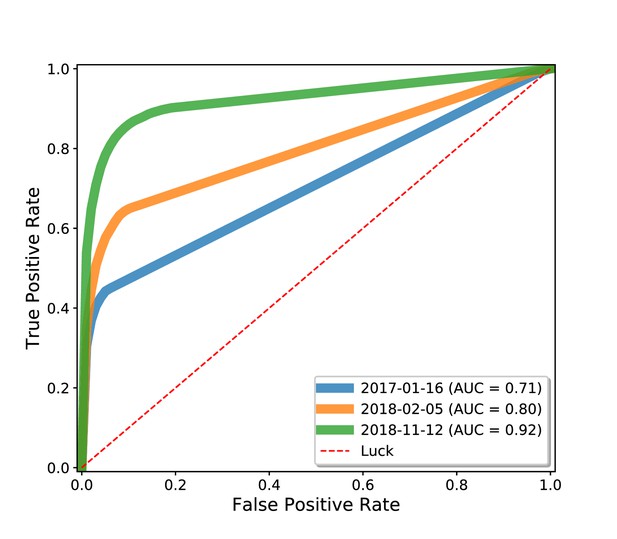
Drug repurposing using the Wikidata knowledge graph.
We analyzed three snapshots of Wikidata using Rephetio, a graph-based algorithm for predicting drug repurposing candidates (Himmelstein et al., 2017). We evaluated the performance of the Rephetio algorithm on three historical versions of the Wikidata knowledge graph, quantified based on the area under the receiver operator characteristic curve (AUC). This analysis demonstrated that the performance of Rephetio in drug repurposing improved over time based only on improvements to the underlying knowledge graph. Details of this analysis can be found at https://github.com/SuLab/WD-rephetio-analysis (archived at Mayers and Su, 2020).
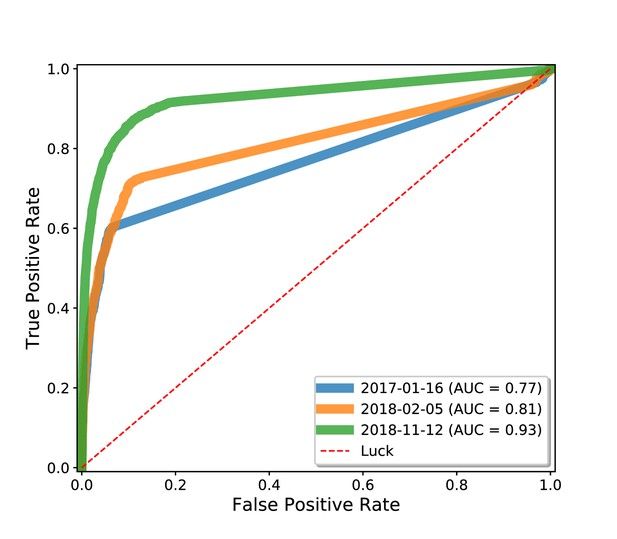
Drug repurposing using the Wikidata knowledge graph, evaluated using an external test set.
The analysis in Figure 5 was based on a cross-validation of indications that were present in Wikidata. This time-resolved analysis was run using an external gold standard set of indications from Drug Central (Ursu et al., 2017).





